HIV GAG Biological Problem
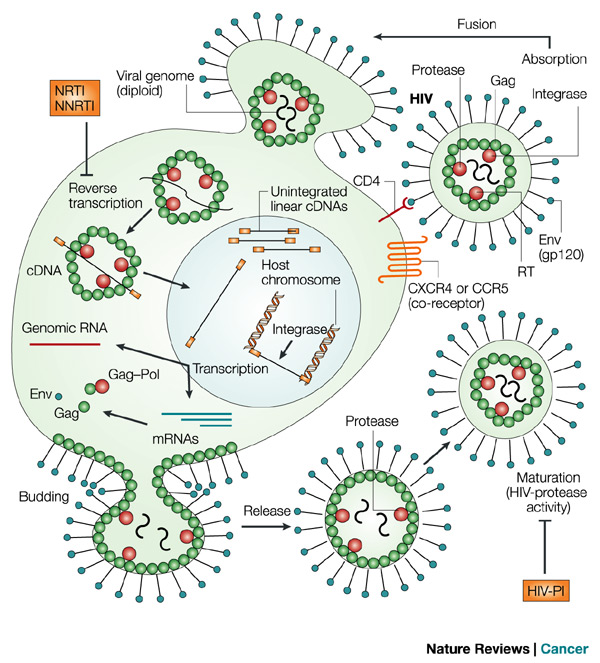
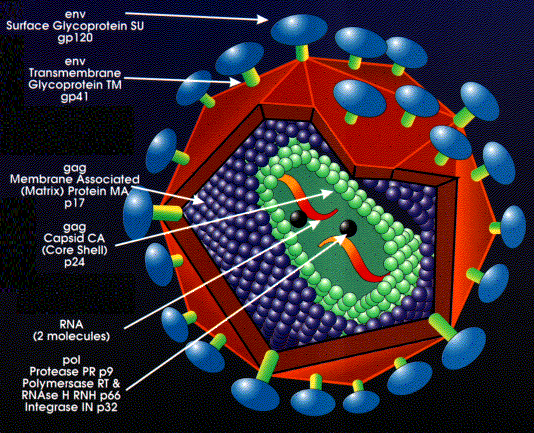
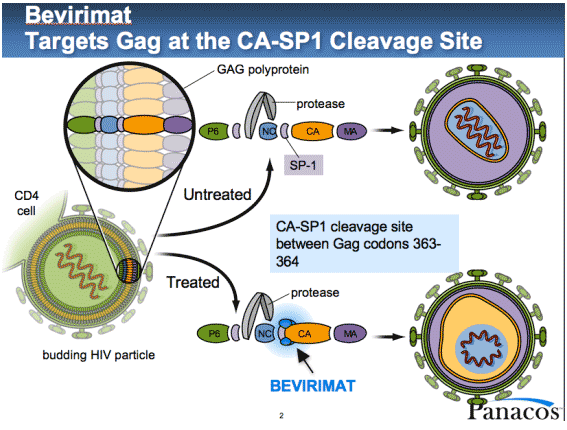 - Emory: The population degenerate consensus for the recipient is not
represented in the consensus for the donor.
- Is there a rare founder in the donor or did the recipient quickly
evolve away ?
- PacBio sequence HIV GAG (1.5kb) donor for 10 transmission pairs:
- Look for evidence of recipient population consensus in reads.
- And multiplex 10,5,4,3 samples on one chip... to keep us on our toes.
Next
================================
- Emory: The population degenerate consensus for the recipient is not
represented in the consensus for the donor.
- Is there a rare founder in the donor or did the recipient quickly
evolve away ?
- PacBio sequence HIV GAG (1.5kb) donor for 10 transmission pairs:
- Look for evidence of recipient population consensus in reads.
- And multiplex 10,5,4,3 samples on one chip... to keep us on our toes.
Next
================================
GAG clustering consensus:
- Cluster consensus samples and a subpopulation might match recipient Sanger estimate. - Clustering plots for 25 runs with single and multiple patients per chip: README_emoryHIVGAG_allclusters.html - Good: multiplexes appear distinshable. Bad: singletons number estimate? - The overall clustering places singleton samples (mostly) together: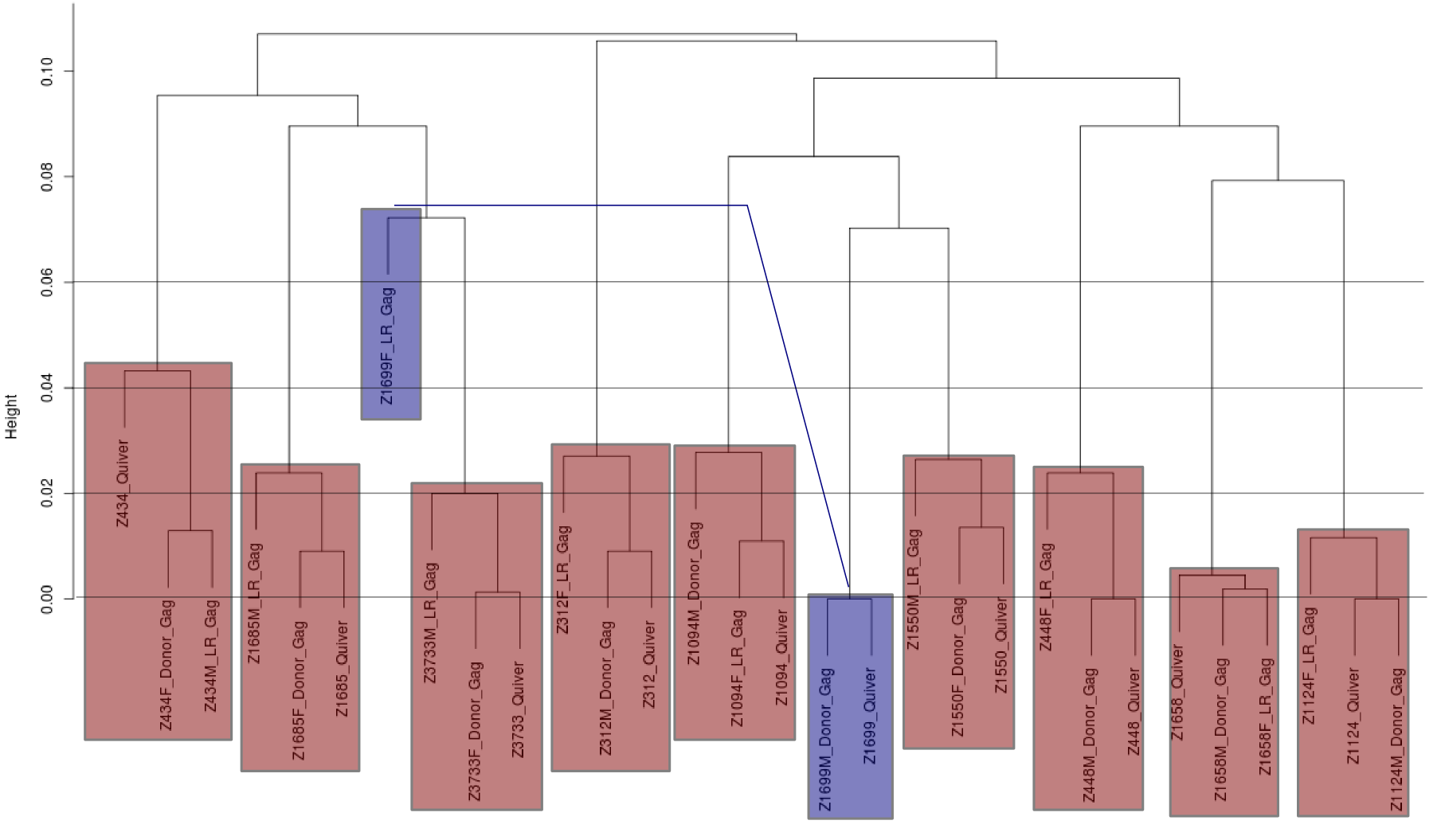 - Sometimes Quiver is closer to receipient, even though donor sample
was sequenced.
- Less than 2% divergence within sample and more than 7% divergent
between samples.
Next
================================
- Sometimes Quiver is closer to receipient, even though donor sample
was sequenced.
- Less than 2% divergence within sample and more than 7% divergent
between samples.
Next
================================
Directed Analysis:
- Search for single best read to recipient in donor sequencing - Score read under recipient Sanger and donor Sanger references. Lower error implies better fit to reference.
+------------------------------------------------------------------------+
|direct_2450423-0001_Z1685 |
| recErr 0.018906 recErrInDonor 0.017556 False 0.106880 |
| |
|direct_2450423-0002_Z1658 |
| recErr 0.026991 recErrInDonor 0.030364 True -0.169883|
| |
|direct_2450423-0017_Z3733 |
| recErr 0.035570 recErrInDonor 0.017450 False 1.027434 |
| |
|direct_2450423-0018_Z448 |
| recErr 0.026104 recErrInDonor 0.048193 True -0.884553|
| |
|direct_2450423-0019_Z1699 |
| recErr 0.098582 recErrInDonor 0.015488 False 2.670173 |
| |
|direct_2450423-0020_Z1094 |
| recErr 0.018996 recErrInDonor 0.095658 True -2.332190|
| |
|direct_2450423-0021_Z1550 |
| recErr 0.029730 recErrInDonor 0.017520 False 0.762917 |
| |
|direct_2450423-0022_Z434 |
| recErr 0.035835 recErrInDonor 0.051247 True -0.516098|
| |
|direct_2450423-0023_Z1124 |
| recErr 0.024324 recErrInDonor 0.041216 True -0.760824|
| |
|direct_2450423-0024_Z312 |
| recErr 0.022927 recErrInDonor 0.051922 True -1.179298|
+------------------------------------------------------------------------+
- Not all cases are clear.
- Looking at -0019 (recipient most likely not present) and
-0024 (evidence of recipient presence)
- For each read score against recipient and the donor and take log
ratio or error.
- Here, a positive score indicates support of the recipient
reference over the donor reference
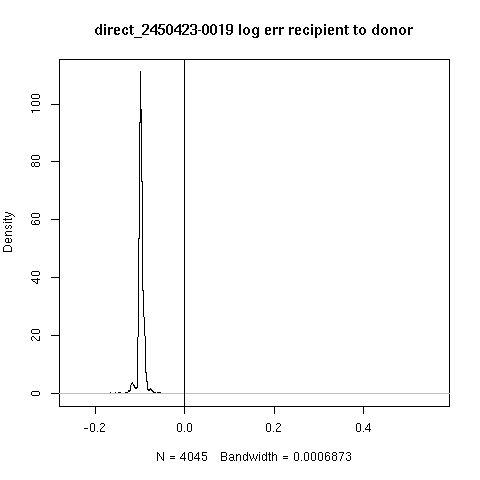
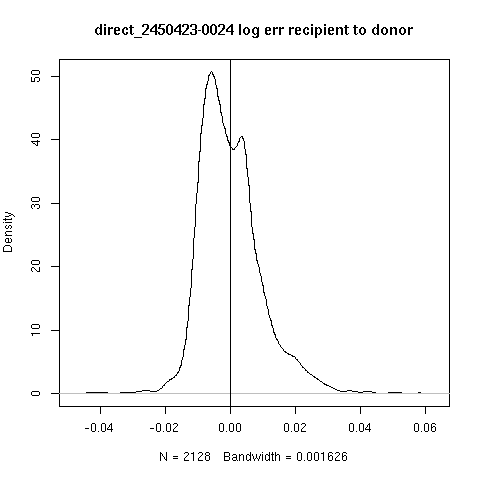 - There appears to be a recipient subpopulation in -0024 but not in -0019.
Next
================================
- There appears to be a recipient subpopulation in -0024 but not in -0019.
Next
================================
Quiver Consensus:
- Estimate Quiver consensus on each run. - Sanity check: singleton Quiver consensus does not agree with Sanger population!
Query: HIVGAG1|quiver
Target: Z1658M_Donor_Gag
Model: affine:local:dna2dna
Raw score: 7233
Query range: 17 -> 1499
Target range: 0 -> 1479
18 : ATGGGTGCGAGAGCGTCAGTATTAAGCGGGGGAAAATTAGACTCATGGGAAAAAATTAGGTT : 79
||||||||||||||||||:|||||||||||||||||||||||||||||||||||||||||||
1 : ATGGGTGCGAGAGCGTCARTATTAAGCGGGGGAAAATTAGACTCATGGGAAAAAATTAGGTT : 62
80 : AAGGCCAGGGGGAAAGAAACACTATATGATGAAACATTTAGTATGGGCAAGCAGGGAGCTGG : 141
|||||||||||||||||||||||||||||||||||||:||||||||||||||||||||||||
63 : AAGGCCAGGGGGAAAGAAACACTATATGATGAAACATYTAGTATGGGCAAGCAGGGAGCTGG : 124
142 : GAAGATTTGCACTTAACCCTGGCCTTTTAGAAACACCAGAAGGCTGTAAACAAATAATGAAA : 203
:||||||||||||||||||||||||||||||:|||:||||||||||||||||||||||::||
125 : RAAGATTTGCACTTAACCCTGGCCTTTTAGARACAYCAGAAGGCTGTAAACAAATAATRMAA : 186
204 : CAGCTGCAACCAGCTCTTCAGACAGGAACAGAGGAACTTAAATCATTATATAACACAATAGC : 265
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
187 : CAGCTGCAACCAGCTCTTCAGACAGGAACAGAGGAACTTAAATCATTATATAACACAATAGC : 248
266 : AACTCTCTATTGTGTACATAAAGGGATAAAGGTACAAGACACCAAGGAAGCCTTAGACAAGA : 327
||||||||||||||||||||||||:|||||||||||||||||||||||||||||||||||||
249 : AACTCTCTATTGTGTACATAAAGGRATAAAGGTACAAGACACCAAGGAAGCCTTAGACAAGA : 310
328 : TAGAGGAAGAACAAAACAAAAGCCAGCAAGGAACACAGCAGGCAAAAGCGGCTGACGAAAAG : 389
|||||||||||||||||||||||||||||::|||||||||||||||||||||||||||||||
311 : TAGAGGAAGAACAAAACAAAAGCCAGCAARRAACACAGCAGGCAAAAGCGGCTGACGAAAAG : 372
390 : GTCAGTCAAAATTATCCTATAGTGCAAAATCAACAAGGACAAATGGTACACCAGGCCATATC : 451
||||||||||||||||||||||||||:|||||:|||||||||||||||||||||||||||||
373 : GTCAGTCAAAATTATCCTATAGTGCARAATCAMCAAGGACAAATGGTACACCAGGCCATATC : 434
452 : ACCTAGAACTTTGAATGCATGGGTAAAAGTGATAGAAGAAAAGGCTTTTAGCCCAGAGGTAA : 513
|||||||||||||||||||||||||||:||||||||||||||||||||||||||||||||||
435 : ACCTAGAACTTTGAATGCATGGGTAAARGTGATAGAAGAAAAGGCTTTTAGCCCAGAGGTAA : 496
514 : TACCCATGTTTACAGCATTATCAGAAGGAGCCACCCCTCAAGATTTAAACACCATGTTAAAT : 575
||||||||||||||||||||||||||||||||||||||||:|||||||||||||||||||||
497 : TACCCATGTTTACAGCATTATCAGAAGGAGCCACCCCTCARGATTTAAACACCATGTTAAAT : 558
576 : ACAGTGGGGGGACATCAAGCAGCCATGCAAATGTTAAAAGATACCATTAATGATGAGGCTGC : 637
|||||||||||||||||||||||||||||||||||||||||||||||:||||||||||||||
559 : ACAGTGGGGGGACATCAAGCAGCCATGCAAATGTTAAAAGATACCATYAATGATGAGGCTGC : 620
638 : AGAATGGGATAGATTACATCCAGTACATGCAGGGCCTATTGCACCAGGCCAAATGAGAGAAC : 699
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
621 : AGAATGGGATAGATTACATCCAGTACATGCAGGGCCTATTGCACCAGGCCAAATGAGAGAAC : 682
700 : CAAGGGGAAGTGACATAGCAGGAACTACTAGTACCCTTCAGGAACAAATAGCATGGATGACA : 761
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
683 : CAAGGGGAAGTGACATAGCAGGAACTACTAGTACCCTTCAGGAACAAATAGCATGGATGACA : 744
762 : AATAACCCACCCATTCCAGTGGGAGAAATATATAAAAGATGGATAATTCTGGGATTAAATAA : 823
|||||:|||||||||||||||||||||||||||||||||||||||||||||||||||||:||
745 : AATAAYCCACCCATTCCAGTGGGAGAAATATATAAAAGATGGATAATTCTGGGATTAAAYAA : 806
824 : AATAGTAAGAATGTATAGCCCTGTCAGCATTTTGGACATAAAACAGGGGCCAAAGGAACCCT : 885
|||||||||||||||||||||||||||||||||||||||||||||:||||||||||||||||
807 : AATAGTAAGAATGTATAGCCCTGTCAGCATTTTGGACATAAAACARGGGCCAAAGGAACCCT : 868
886 : TTAGAGATTATGTAGACCGGTTCTTTAAAACTTTAAGAGCTGAACAAGCTACACAAGAAGTA : 947
|||||||:||||||||:|||||||||||||||||||||||||||||||||||||||||||||
869 : TTAGAGAYTATGTAGAYCGGTTCTTTAAAACTTTAAGAGCTGAACAAGCTACACAAGAAGTA : 930
948 : AAAAATTGGATGACAGACACCTTGCTGGTCCAAAATGCAAACCCAGATTGTAAGTCCATTTT : 1009
|||::|||||||||||||||:||||||:||||||||||||||||||||||||||::||||||
931 : AAARRTTGGATGACAGACACMTTGCTGRTCCAAAATGCAAACCCAGATTGTAAGWSCATTTT : 992
1010 : AAAAGCATTAGGATCAGGGGCTTCATTAGAAGAAATGATGACAGCATGTCAAGGAGTGGGAG : 1071
|||||||||||||||||||||| ||||||||||||||||||||||||||||:||||||||||
993 : AAAAGCATTAGGATCAGGGGCTSCATTAGAAGAAATGATGACAGCATGTCARGGAGTGGGAG : 1054
1072 : GACCTAGCCACAAAGCAAGAGTATTGGCTGAGGCAATGAGCCAAGCACAAAGTACAAACATA : 1133
||||||||||||||||||||||:|||||||||||||||||||||||||| ||||||||||||
1055 : GACCTAGCCACAAAGCAAGAGTRTTGGCTGAGGCAATGAGCCAAGCACACAGTACAAACATA : 1116
1134 : CTGATGCAGAGAAGCAATTTTAAAGGCCCTAAAAGAATAGTTAAATGTTTCAATTGTGGCAA : 1195
||||||||||||||||||||||||||||||||||||||:|||||||||||||||||||||||
1117 : CTGATGCAGAGAAGCAATTTTAAAGGCCCTAAAAGAATWGTTAAATGTTTCAATTGTGGCAA : 1178
1196 : AGAAGGGCACATAGCCAGAAATTGCAGGGCCCCTAGGAAAAAGGGCTGTTGGAAATGTGGAA : 1257
:|||||||||||||||||||||||||||||||||||||||||||||||||||||:|||||||
1179 : RGAAGGGCACATAGCCAGAAATTGCAGGGCCCCTAGGAAAAAGGGCTGTTGGAARTGTGGAA : 1240
1258 : AGGAAGGACACCAAATGAAAGACTGTAATAATGAGAGACAGGCCAATTTTTTAGGGAGAATT : 1319
||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||
1241 : AGGAAGGACACCAAATGAAAGACTGTAAT---GAGAGACAGGCCAATTTTTTAGGGAGAATT : 1299
1320 : TGGCCTTCCCACAAGGGGAGGCCAGGAAATTTCCTTCAGAGCAGGCCAGAGCCGACAGCTCC : 1381
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1300 : TGGCCTTCCCACAAGGGGAGGCCAGGAAATTTCCTTCAGAGCAGGCCAGAGCCGACAGCTCC : 1361
1382 : ACCAGCAGAGAGCTTCAGGTTCGAGGAAACAACCCCTGCTCCGAAGCAGGAGATGAAGGACA : 1443
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1362 : ACCAGCAGAGAGCTTCAGGTTCGAGGAAACAACCCCTGCTCCGAAGCAGGAGATGAAGGACA : 1423
1444 : GGGAACCCTTAACTTCCCTCAAATCACTCTTTGGCAGCGACCCCTTGTCTCAATAA : 1499
||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1424 : GGGAACCCTTAACTTCCCTCAAATCACTCTTTGGCAGCGACCCCTTGTCTCAATAA : 1479
Next
================================
Ambiguous Consensus Estimation:
- An error source might be Emory's Sanger population ambiguities. - Under the quiver consensus, there are many mixed columns.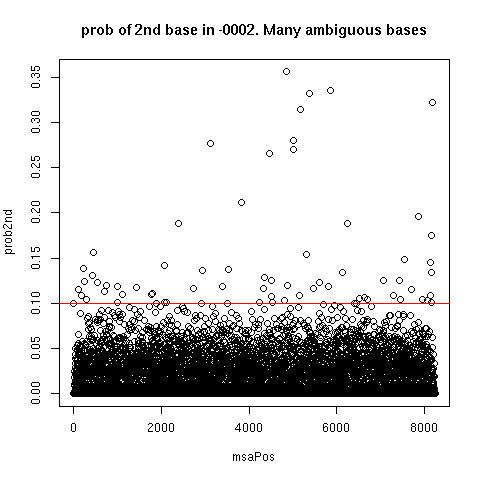
 - Estimate ambiguous consensus from Quiver with minor frequency
threshold.
Next
================================
- Estimate ambiguous consensus from Quiver with minor frequency
threshold.
Next
================================
Shifting Degenerate Consensus:
- Vary degenerate threshold and align to Sanger donor and recipient consensus (run -0002).
+-------+--------+
| Thresh|BestHit |
|-------+--------+
| 0.1 |recip |
| 0.3 |recip |
| 0.35 |recip |
| 0.38 |donor |
| 0.4 |donor |
| 0.5 |donor |
+-------+--------+
- Depending on ambiguity threshold, the alignment switches between
recipient and donor (with addition of 3 ambiguities)!
- The alignments with ambiguous bases:
degenerate shift alignment
- Can anyone trust population Sanger?
Next
================================
One Billion Genomes?:
- 30 ambiguous bases at 10% in singleton run -0002. - ~1 billion (=2^30) unique genomes if independent.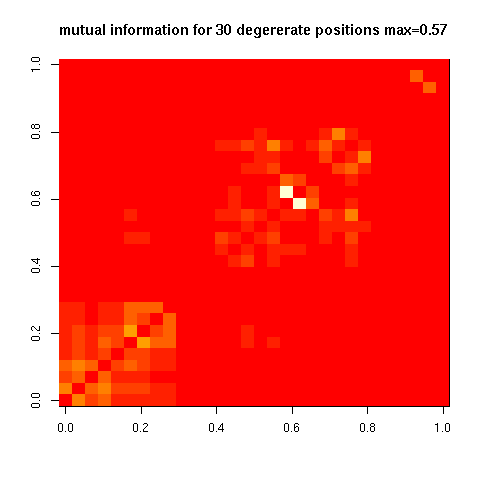 - Most positions are largely independent.
- Here is the highest mutual information:
- Most positions are largely independent.
- Here is the highest mutual information:
+------------------------------+
| [,A] [,C] [,G] [,T] [,-] |
|[A,] 64 286 1736 0 2 |
|[C,] 1 83 0 0 0 |
|[G,] 108 370 202 0 3 |
|[T,] 17 3666 58 30 208 |
|[-,] 59 417 24 0 130 |
+------------------------------+
- Possible haplotypes around these two adjacent positions are:
AGTCCA, AGAGCA
- -0002 has 20 selected variant features (missing 10 from the 30 at
10% but largely agree). This gives 0.8 (lenient 0.425) threshold.
 - Are we sequencing 1 billion genomes in this sample?
- Are we sequencing 1 billion genomes in this sample?