goal: results of preliminary look at HIV sequencing
- Examine sequence data and references from two patients with time points and replicates 222 has samples from day 18, 101, and 105. 9213 has samples from day 28, 59, and 165. - Summary: - Second half HIV genomes sequenced (~4.5kb) - Consensus genomes estimated for each of the runs. - SNP variant analysis against references - Mixed populations of ~300-base deletions variants in ENV - Complexity of the sample mixtures changes between time points. Next ================================Inputs and Strategy
- Here are the references and data I got:
Reference Data
X /222-101/C01_1/Analysis_Results/
X /222-101_15Mar/C01_1/Analysis_Results/
X /222-101_15Mar/D01_1/Analysis_Results/
X X /222-101_14Mar -> NO DATA!
AC222_D18_assembly.fa /222-18_14Mar/A01_1/Analysis_Results/
AC222_D18_assembly.fa /222-18_15Mar/A01_1/Analysis_Results/
AC222_D18_assembly.fa /222-18_15Mar/B01_1/Analysis_Results/
AC222_D105_assembly.fa /222-105_14Mar/B01_1/Analysis_Results/
AC222_D105_assembly.fa /222-105_15Mar/C01_1/Analysis_Results/
9213_D0.fa X
9213_D28.fa /9213-28_14Mar/E01_1/Analysis_Results/
9213_D28.fa /9213-28_15Mar/F01_1/Analysis_Results/
9213_D28.fa /9213-28_15Mar/G01_1/Analysis_Results/
9213_D28.fa /9213-28_26Apr/D01_1/Analysis_Results/
9213_D28.fa /9213-28_26Apr/D01_1/Analysis_Results/
9213_D59.fa /9213-59_14Mar/D01_2/Analysis_Results/
9213_D59.fa /9213-59_15Mar/E01_1/Analysis_Results/
9213_D165.fa /9213-165_14Mar/F01_1/Analysis_Results/
9213_D165.fa /9213-165_15Mar/H01_1/Analysis_Results/
X = missing
- Strategy:
- Clinical samples can present a complex mixture of HIV genomes.
- Rely on PacBio long reads to sequence entire genomes as single
molecules.
- Complete genome characterizations where different subspecies
present in mixtures can be separated out.
Next
================================
First look at P222-D105
- Examine 222-105-15Mar against given reference to get an idea of what is being sequenced. - Longest mapped read is ~5kb
Mapped readlengths:
Min. 1st Qu. Median Mean 3rd Qu. Max.
21 494 1158 1344 1988 5226
- Distribution of mapped read lengths and log coverage across given
reference genome
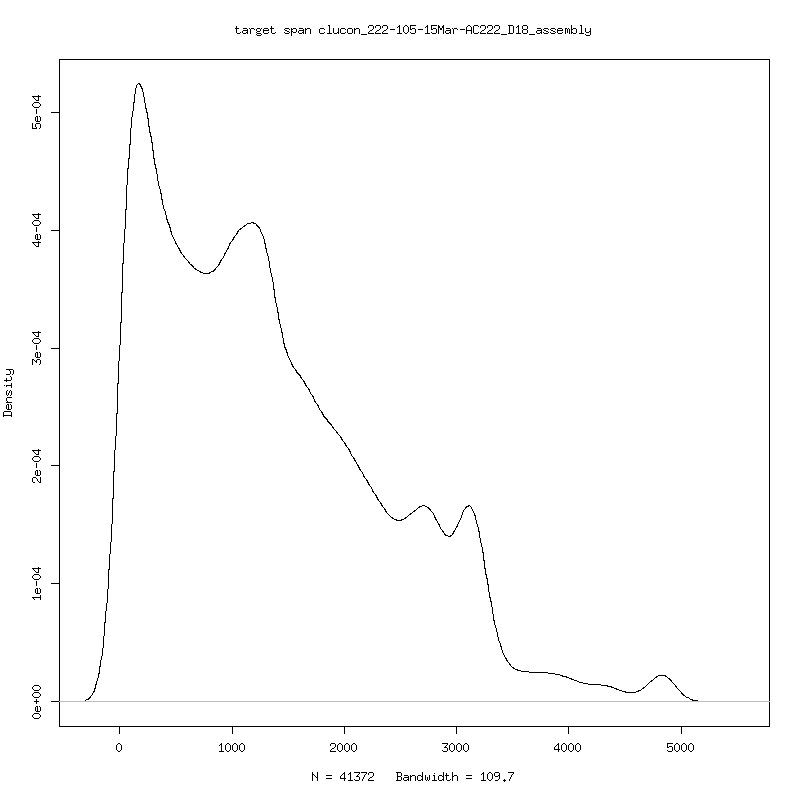
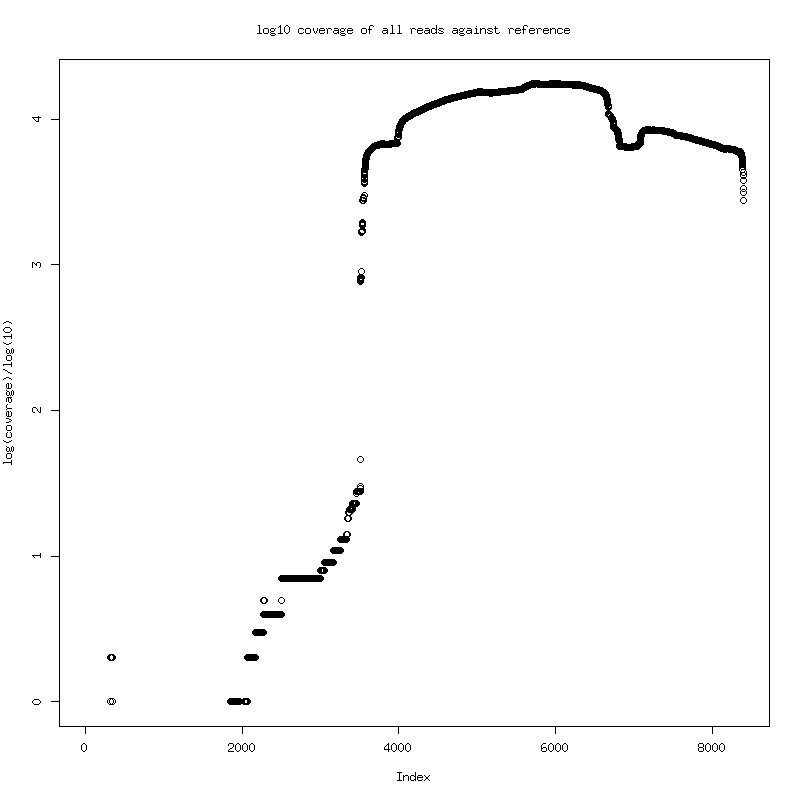 - Appears to be second half HIV genome sequencing. Trim reference to the highly
covered region (4656 bases) for some analysis
Next
================================
- Appears to be second half HIV genome sequencing. Trim reference to the highly
covered region (4656 bases) for some analysis
Next
================================
Cluster consensus P222-D105
- Compare all 4kb reads against each other. There appear to be at least 3 major groups with fractions 44%, 36% and 20%: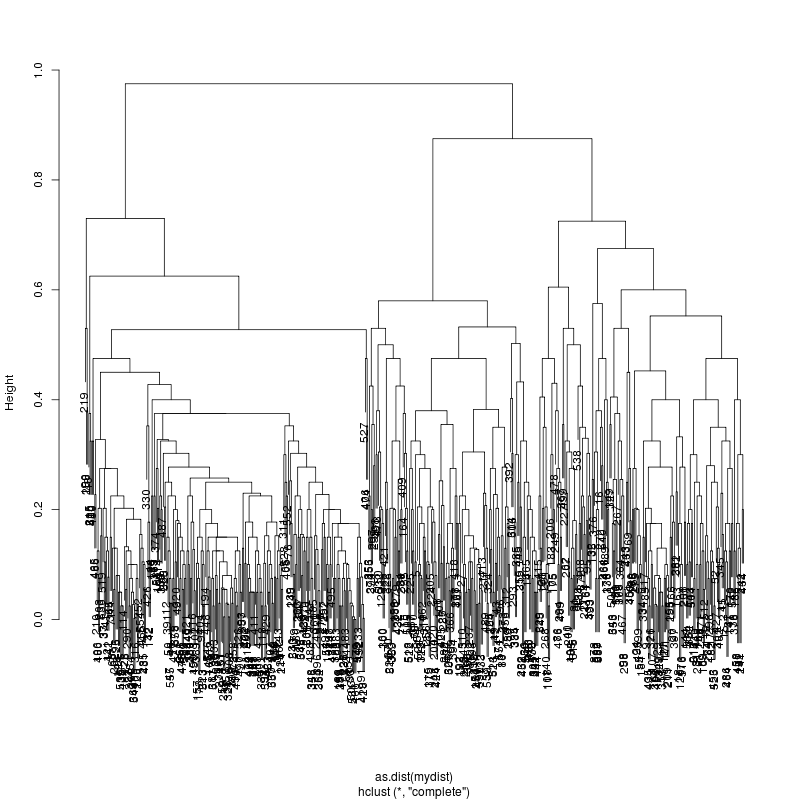 - Estimate consensus within each of the three subgroups and show
pairwise alignments:
blasr-222-105_15Mar.12.output
blasr-222-105_15Mar.13.output
blasr-222-105_15Mar.23.output
- Sample 222-105-15Mar has a large ~300 base delete variant at 44%
abundance. There also appear to be two codon differences in the second
and third most abundant subspecies.
Next
================================
- Estimate consensus within each of the three subgroups and show
pairwise alignments:
blasr-222-105_15Mar.12.output
blasr-222-105_15Mar.13.output
blasr-222-105_15Mar.23.output
- Sample 222-105-15Mar has a large ~300 base delete variant at 44%
abundance. There also appear to be two codon differences in the second
and third most abundant subspecies.
Next
================================
Delete variant P222-D105
- Blast subgroup consensus sequences against NR database ---- Majority subgroup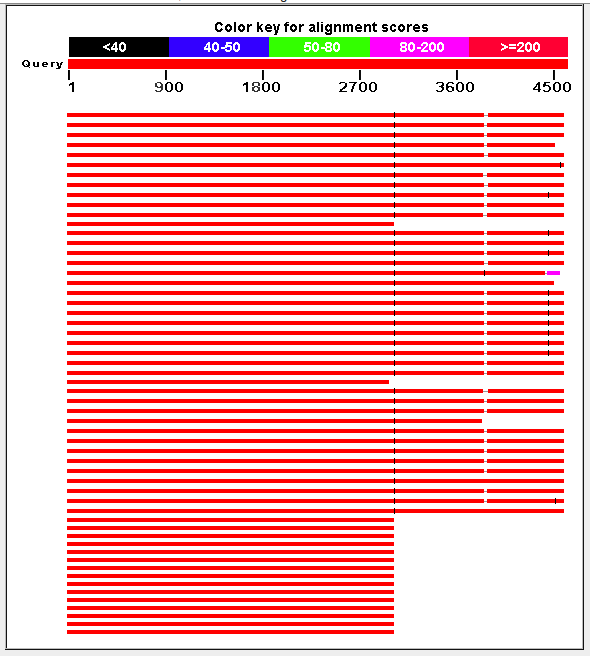 Staggered hits with "unaligned" gaps inbetween:
Staggered hits with "unaligned" gaps inbetween:
Hit "HIV-1 isolate 5082-86 clone pbf26 from USA, complete genome"
query database
1:3023 4974:7996
- 0 gap 42 gap
3023:3858 8039:8876
- 50 gap 2 gap
3898:4593 8880:9572
4121:4593 1:473 ??? end maps to begining duplicated. 1:473 like
9099:9571 in reference ???
---- Second most abundant subgroup
Same database hit
query database
1:2735 4974:7708
- -5 332
2728:3566 8038:8876
- 8 2
3606:4301 8880:9572
3829:4301 1:473
- Missing about 300 bases in the query between segment 1 and 2
(reference bases 7708-7996)...
- The deletion appears to occurs in ENV (at the end of GP120 into the
heptad repeat. "heptad repeat 1-heptad repeat 2 region (ectodomain)
of the gp41, HR1 Site, homotrimer interface [polypeptide binding]")
- Google shows "Impact of the HIV-1 env Genetic Context outside
HR1-HR2 on Resistance to the Fusion Inhibitor Enfuvirtide and Viral
Infectivity in Clinical Isolates"
Next
================================
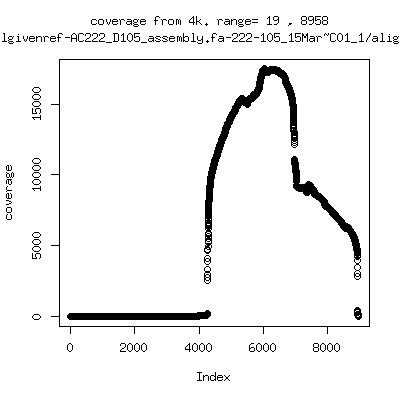
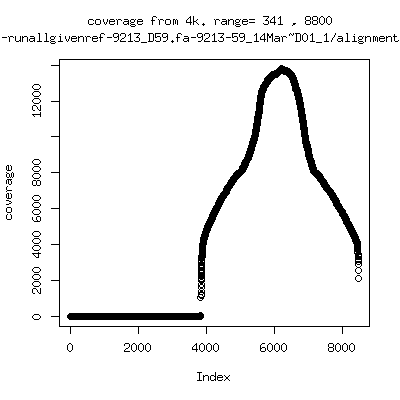
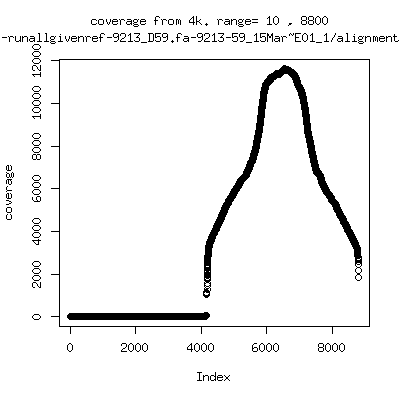
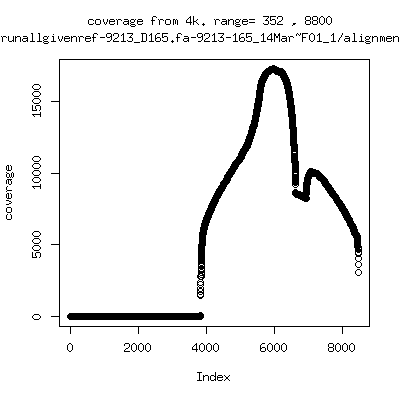
Coverage all runs against their references
- Coverage plots of all reads against given references. Note P222-101 not shown because no reference given. ---- P222-D18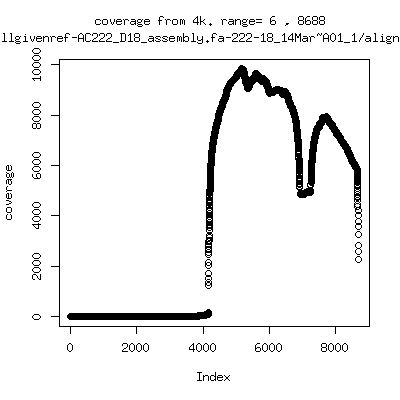
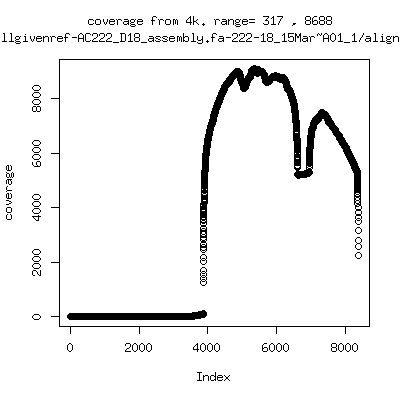
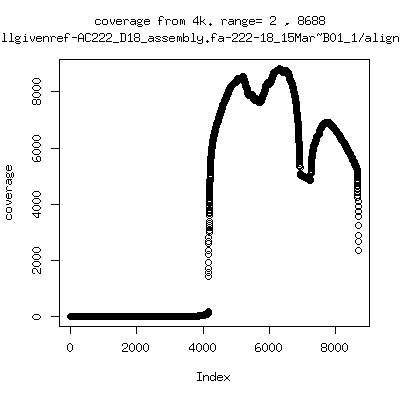 ---- P222-D105
---- P222-D105
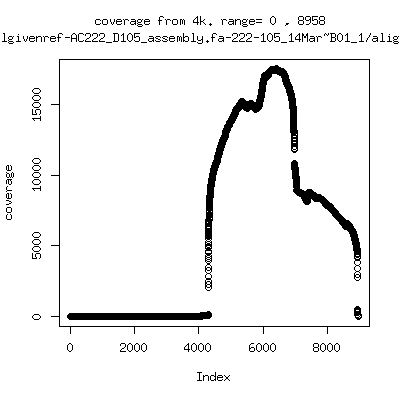
 ---- P9213-D59
---- P9213-D59
 ---- P9213-D165
---- P9213-D165
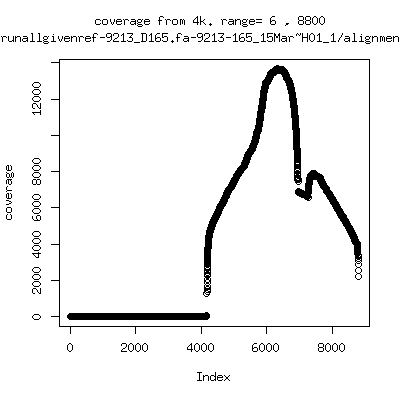 ---- P9213-D28: NO mapped reads > 4kb (max = 2460)
---- P9213-D28: NO mapped reads > 4kb (max = 2460)
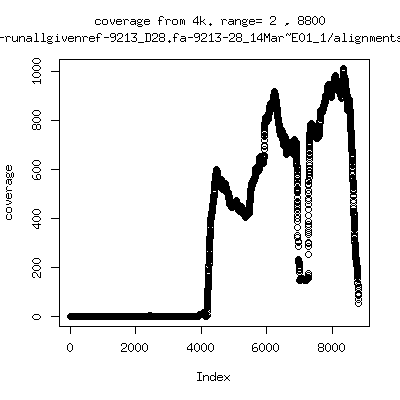
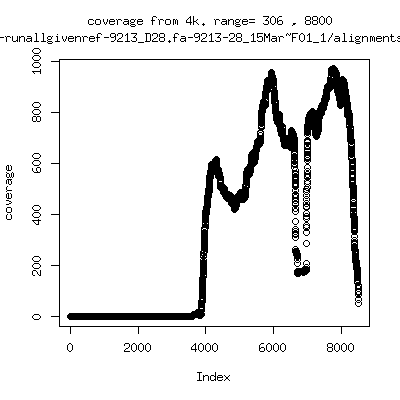
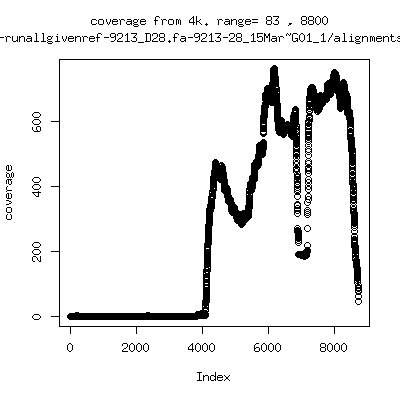
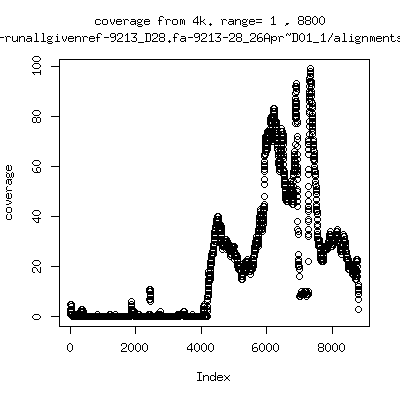
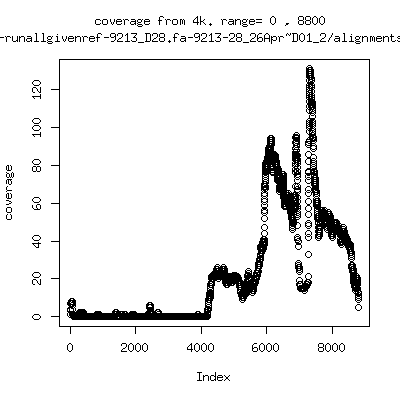 - Interestingly, I can distinguish patients+day by looking at the
all-reads coverage patterns.
Next
================================
- Interestingly, I can distinguish patients+day by looking at the
all-reads coverage patterns.
Next
================================
Minor variant p-value all runs against given references
- Align reads against given reference looking ONLY at >4kb maps. - For each column compute p-value of whether that columns might contain a minor variant. ---- P222-D18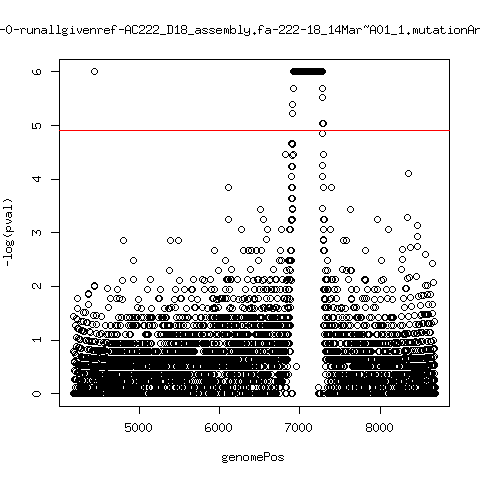
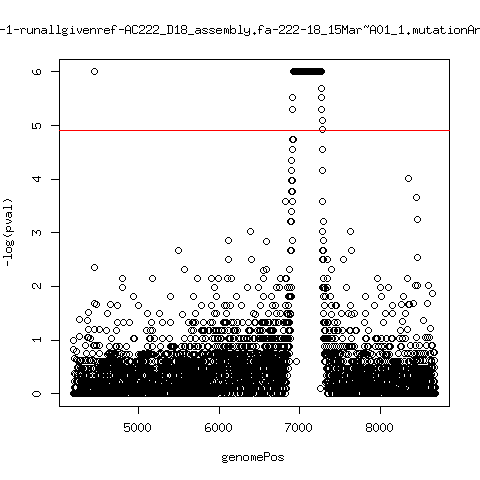
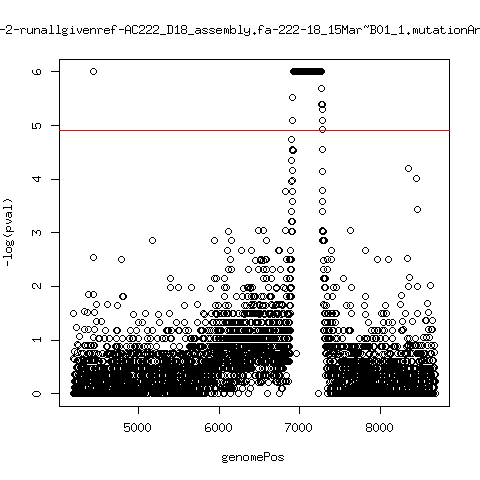 ---- P222-D105
---- P222-D105
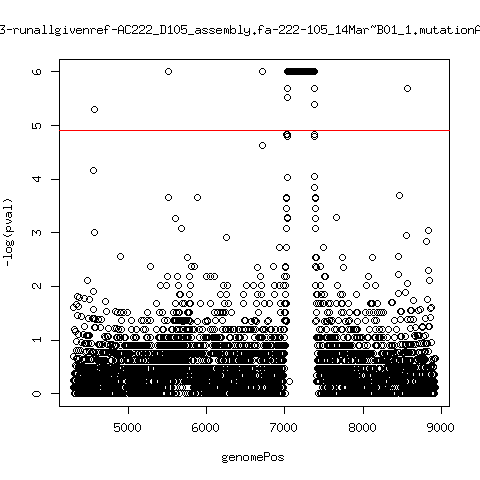
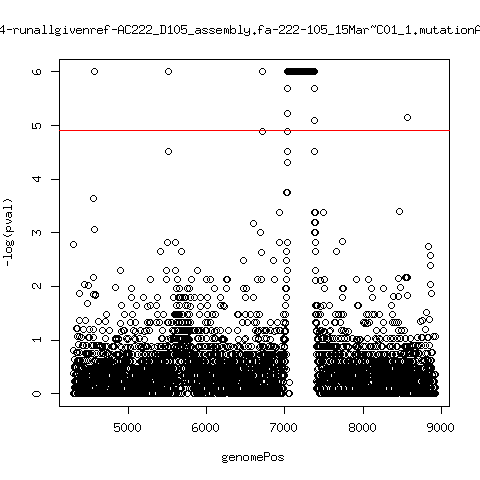 ---- P9213-D59
---- P9213-D59
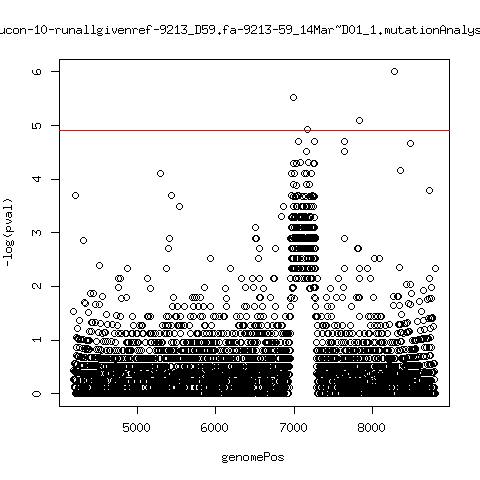
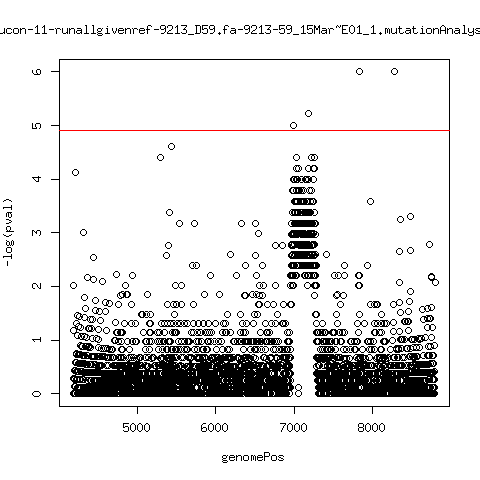 ---- P9213-D165
---- P9213-D165
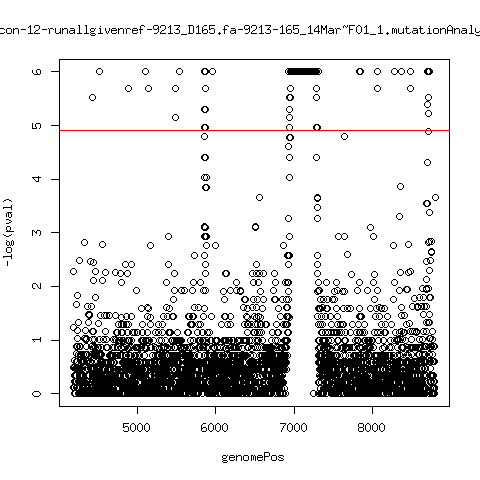
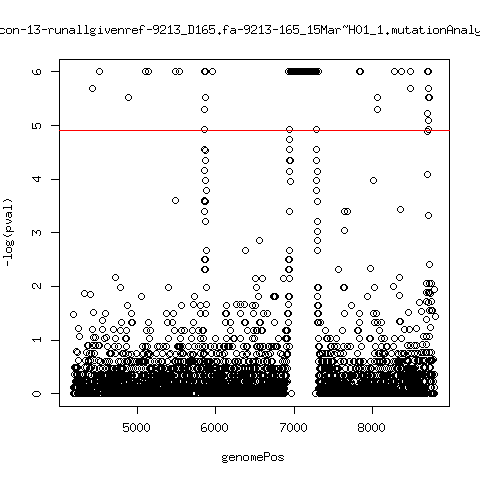 - List of top p-value computations:
---- P222-D18
ma_clucon-0-runallgivenref-AC222_D18_assembly.fa-222-18_14Mar~A01_1.mutationAnalysis.output.top
ma_clucon-1-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~A01_1.mutationAnalysis.output.top
ma_clucon-2-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~B01_1.mutationAnalysis.output.top
---- P222-D105
ma_clucon-3-runallgivenref-AC222_D105_assembly.fa-222-105_14Mar~B01_1.mutationAnalysis.output.top
ma_clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.mutationAnalysis.output.top
---- P9213-D59
ma_clucon-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.mutationAnalysis.output.top
ma_clucon-11-runallgivenref-9213_D59.fa-9213-59_15Mar~E01_1.mutationAnalysis.output.top
---- P9213-D165
ma_clucon-12-runallgivenref-9213_D165.fa-9213-165_14Mar~F01_1.mutationAnalysis.output.top
ma_clucon-13-runallgivenref-9213_D165.fa-9213-165_15Mar~H01_1.mutationAnalysis.output.top
Next
================================
- List of top p-value computations:
---- P222-D18
ma_clucon-0-runallgivenref-AC222_D18_assembly.fa-222-18_14Mar~A01_1.mutationAnalysis.output.top
ma_clucon-1-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~A01_1.mutationAnalysis.output.top
ma_clucon-2-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~B01_1.mutationAnalysis.output.top
---- P222-D105
ma_clucon-3-runallgivenref-AC222_D105_assembly.fa-222-105_14Mar~B01_1.mutationAnalysis.output.top
ma_clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.mutationAnalysis.output.top
---- P9213-D59
ma_clucon-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.mutationAnalysis.output.top
ma_clucon-11-runallgivenref-9213_D59.fa-9213-59_15Mar~E01_1.mutationAnalysis.output.top
---- P9213-D165
ma_clucon-12-runallgivenref-9213_D165.fa-9213-165_14Mar~F01_1.mutationAnalysis.output.top
ma_clucon-13-runallgivenref-9213_D165.fa-9213-165_15Mar~H01_1.mutationAnalysis.output.top
Next
================================
Single consensus all runs
- Single consensus estimates for each of the 4kb runs (the most average sequence in the sample). - Clustering of all consensus sequences shows patients group but days are spread. P222-D101 is distinct (no strong subpopulations)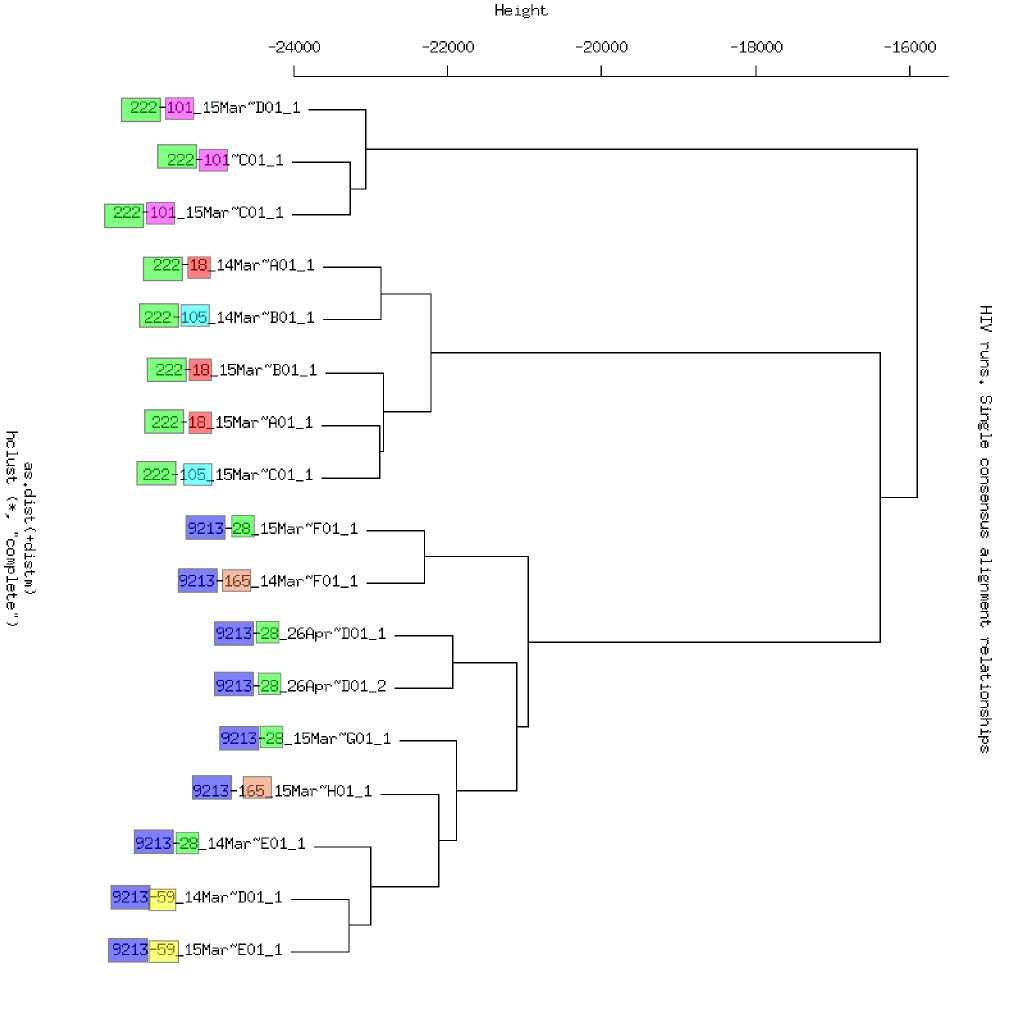 - The single consensus sequences
---- P222-D18
cluconFrom4k-0-runallgivenref-AC222_D18_assembly.fa-222-18_14Mar~A01_1/quiverResult.consensus.fastq
cluconFrom4k-1-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~A01_1/quiverResult.consensus.fastq
cluconFrom4k-2-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~B01_1/quiverResult.consensus.fastq
---- P222-D101
cluconFrom4k-14-runallgivenref-X-222-101~C01_1/quiverResult.consensus.fastq
cluconFrom4k-15-runallgivenref-X-222-101_15Mar~C01_1/quiverResult.consensus.fastq
cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1/quiverResult.consensus.fastq
---- P222-D105
cluconFrom4k-3-runallgivenref-AC222_D105_assembly.fa-222-105_14Mar~B01_1/quiverResult.consensus.fastq
cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/quiverResult.consensus.fastq
---- P9213-D28
cluconFrom4k-5-runallgivenref-9213_D28.fa-9213-28_14Mar~E01_1/quiverResult.consensus.fastq
cluconFrom4k-6-runallgivenref-9213_D28.fa-9213-28_15Mar~F01_1/quiverResult.consensus.fastq
cluconFrom4k-7-runallgivenref-9213_D28.fa-9213-28_15Mar~G01_1/quiverResult.consensus.fastq
cluconFrom4k-8-runallgivenref-9213_D28.fa-9213-28_26Apr~D01_1/quiverResult.consensus.fastq
cluconFrom4k-9-runallgivenref-9213_D28.fa-9213-28_26Apr~D01_2/quiverResult.consensus.fastq
---- P9213-D59
cluconFrom4k-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1/quiverResult.consensus.fastq
cluconFrom4k-11-runallgivenref-9213_D59.fa-9213-59_15Mar~E01_1/quiverResult.consensus.fastq
---- P9213-D165
cluconFrom4k-12-runallgivenref-9213_D165.fa-9213-165_14Mar~F01_1/quiverResult.consensus.fastq
cluconFrom4k-13-runallgivenref-9213_D165.fa-9213-165_15Mar~H01_1/quiverResult.consensus.fastq
Next
================================
- The single consensus sequences
---- P222-D18
cluconFrom4k-0-runallgivenref-AC222_D18_assembly.fa-222-18_14Mar~A01_1/quiverResult.consensus.fastq
cluconFrom4k-1-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~A01_1/quiverResult.consensus.fastq
cluconFrom4k-2-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~B01_1/quiverResult.consensus.fastq
---- P222-D101
cluconFrom4k-14-runallgivenref-X-222-101~C01_1/quiverResult.consensus.fastq
cluconFrom4k-15-runallgivenref-X-222-101_15Mar~C01_1/quiverResult.consensus.fastq
cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1/quiverResult.consensus.fastq
---- P222-D105
cluconFrom4k-3-runallgivenref-AC222_D105_assembly.fa-222-105_14Mar~B01_1/quiverResult.consensus.fastq
cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/quiverResult.consensus.fastq
---- P9213-D28
cluconFrom4k-5-runallgivenref-9213_D28.fa-9213-28_14Mar~E01_1/quiverResult.consensus.fastq
cluconFrom4k-6-runallgivenref-9213_D28.fa-9213-28_15Mar~F01_1/quiverResult.consensus.fastq
cluconFrom4k-7-runallgivenref-9213_D28.fa-9213-28_15Mar~G01_1/quiverResult.consensus.fastq
cluconFrom4k-8-runallgivenref-9213_D28.fa-9213-28_26Apr~D01_1/quiverResult.consensus.fastq
cluconFrom4k-9-runallgivenref-9213_D28.fa-9213-28_26Apr~D01_2/quiverResult.consensus.fastq
---- P9213-D59
cluconFrom4k-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1/quiverResult.consensus.fastq
cluconFrom4k-11-runallgivenref-9213_D59.fa-9213-59_15Mar~E01_1/quiverResult.consensus.fastq
---- P9213-D165
cluconFrom4k-12-runallgivenref-9213_D165.fa-9213-165_14Mar~F01_1/quiverResult.consensus.fastq
cluconFrom4k-13-runallgivenref-9213_D165.fa-9213-165_15Mar~H01_1/quiverResult.consensus.fastq
Next
================================
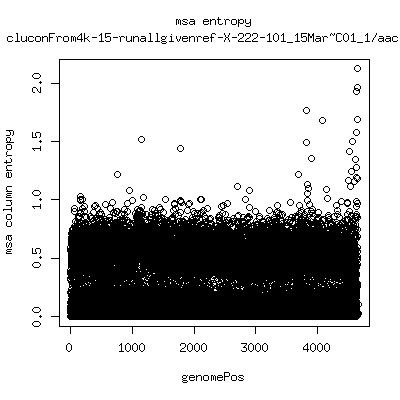
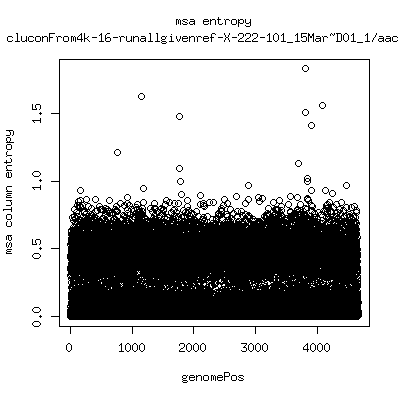
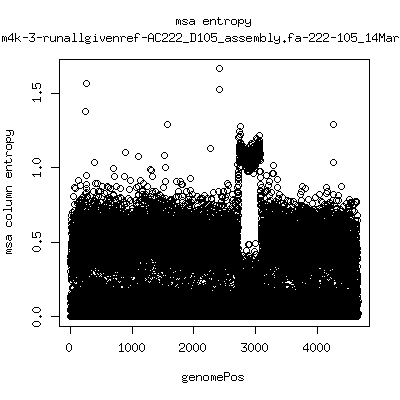
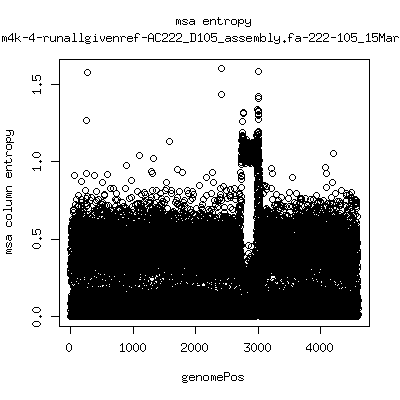
Alignment entropy all runs
- Start with trimmed ~4.5kb reference from P222-D105 as initial seed reference. - Estimate new reference based on >4kb mapped reads. - Compute alignment entropy which measures possibility of mixtures of differing subpopulations present in sample. (P9213_D28 not applicable because of short mapped reads): ---- P222-D18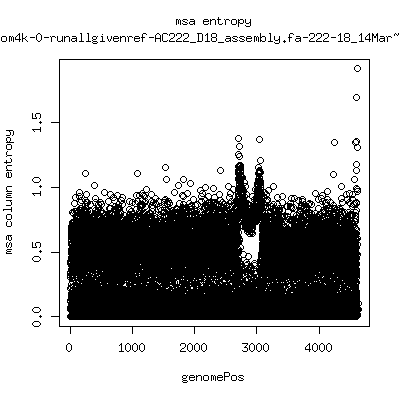
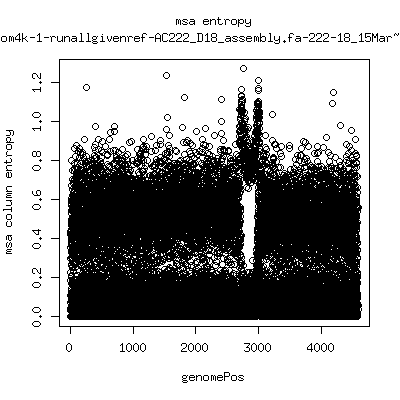
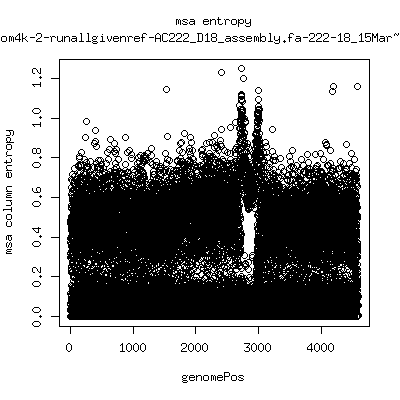 ---- P222-D101 (that had no given reference)
---- P222-D101 (that had no given reference)
 ---- P222-D105
---- P222-D105
 ---- P9213-D59
---- P9213-D59
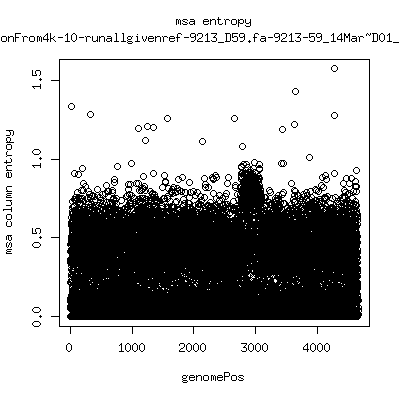
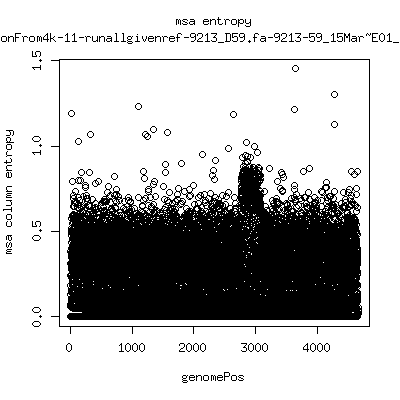 ---- P9213-D165
---- P9213-D165
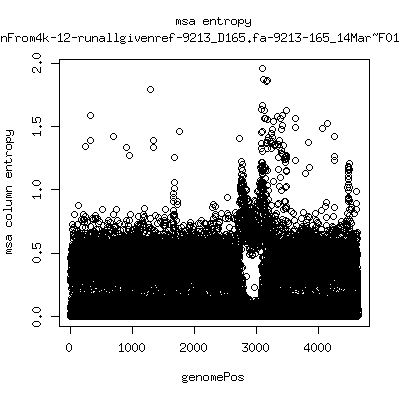
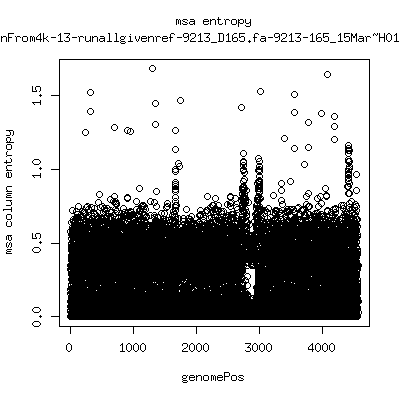 - Patient 222 appears to show varying levels of subpopulations with
low point at day-101. Patient 9213 also shows differences between
day 59 and day 165 (day 28 has low quality).
Next
================================
- Patient 222 appears to show varying levels of subpopulations with
low point at day-101. Patient 9213 also shows differences between
day 59 and day 165 (day 28 has low quality).
Next
================================
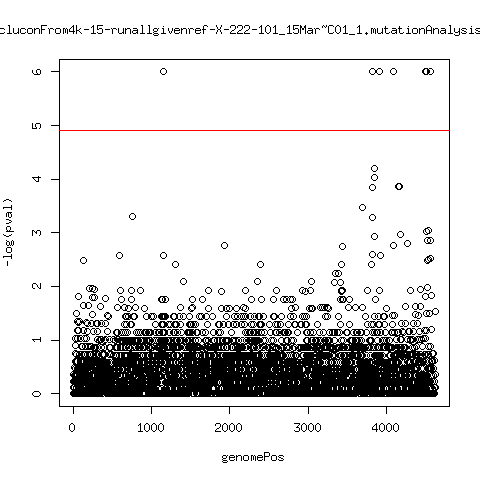
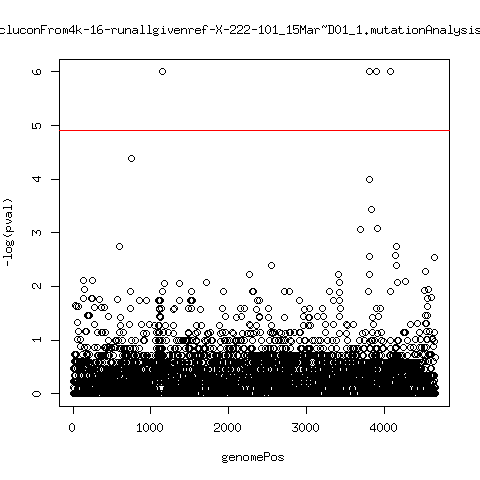
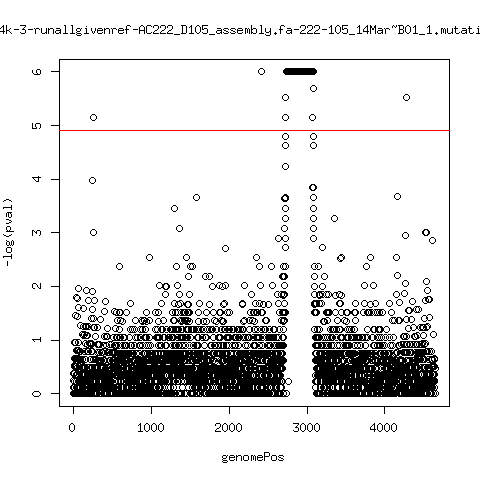
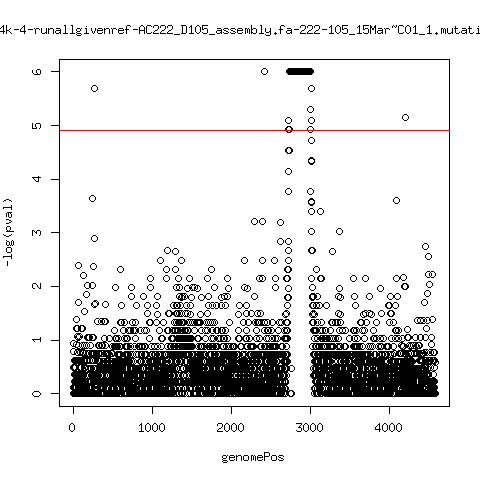
Estimated consensus variant analysis
- For the sample specific consensus, estimate p-values of minor variant ---- P222-D18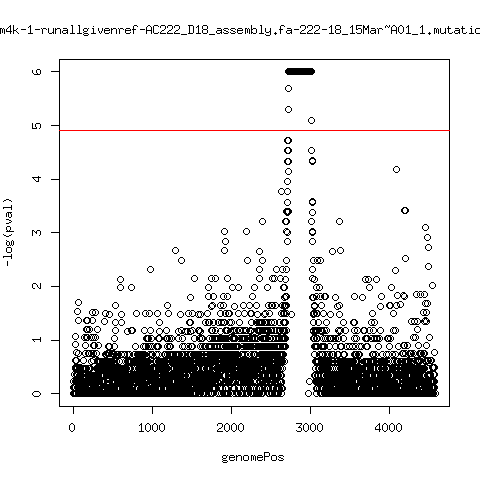
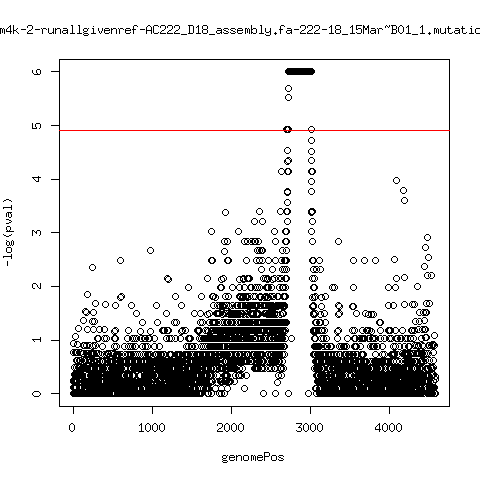
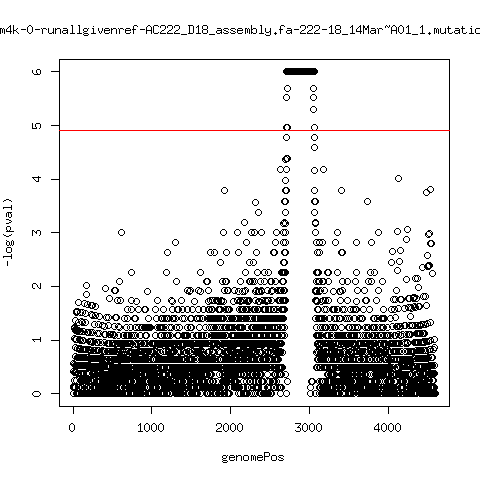 ---- P222-D101
---- P222-D101
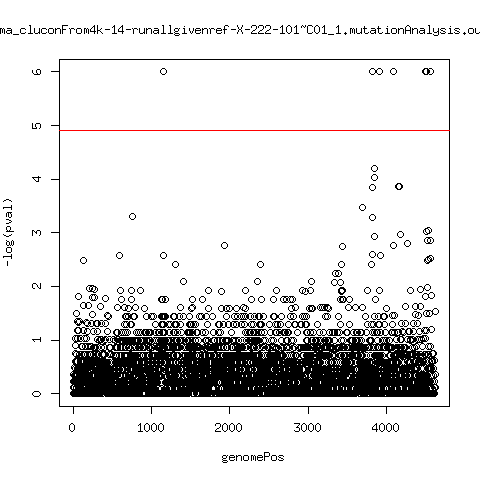
 ---- P222-D105
---- P222-D105
 ---- P9213-D59
---- P9213-D59
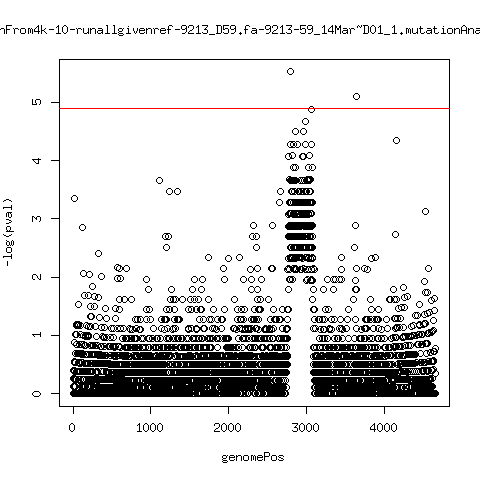
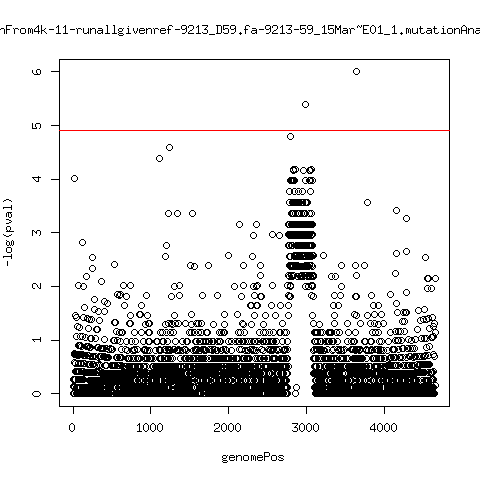 ---- P9213-D165
---- P9213-D165
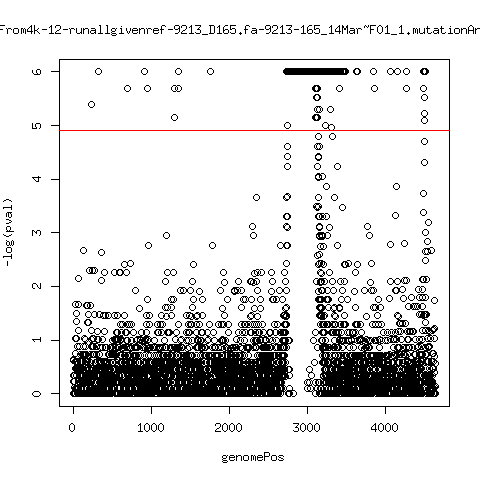
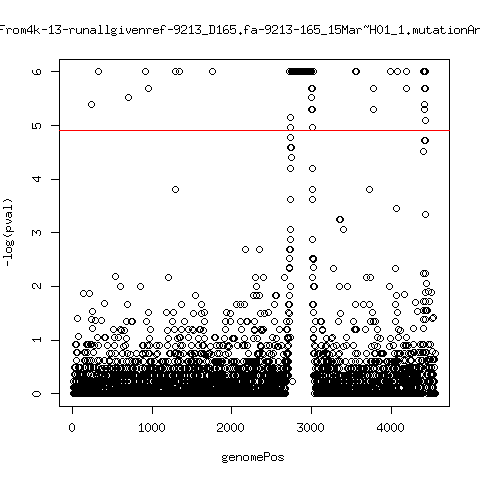 - List of top p-values
---- P222-D18
ma_cluconFrom4k-1-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~A01_1.mutationAnalysis.output.top
ma_cluconFrom4k-2-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~B01_1.mutationAnalysis.output.top
ma_cluconFrom4k-0-runallgivenref-AC222_D18_assembly.fa-222-18_14Mar~A01_1.mutationAnalysis.output.top
---- P222-D101
ma_cluconFrom4k-14-runallgivenref-X-222-101~C01_1.mutationAnalysis.output.top
ma_cluconFrom4k-15-runallgivenref-X-222-101_15Mar~C01_1.mutationAnalysis.output.top
ma_cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1.mutationAnalysis.output.top
---- P222-D105
ma_cluconFrom4k-3-runallgivenref-AC222_D105_assembly.fa-222-105_14Mar~B01_1.mutationAnalysis.output.top
ma_cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.mutationAnalysis.output.top
---- P9213-D59
ma_cluconFrom4k-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.mutationAnalysis.output.top
ma_cluconFrom4k-11-runallgivenref-9213_D59.fa-9213-59_15Mar~E01_1.mutationAnalysis.output.top
---- P9213-D165
ma_cluconFrom4k-12-runallgivenref-9213_D165.fa-9213-165_14Mar~F01_1.mutationAnalysis.output.top
ma_cluconFrom4k-13-runallgivenref-9213_D165.fa-9213-165_15Mar~H01_1.mutationAnalysis.output.top
Next
================================
- List of top p-values
---- P222-D18
ma_cluconFrom4k-1-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~A01_1.mutationAnalysis.output.top
ma_cluconFrom4k-2-runallgivenref-AC222_D18_assembly.fa-222-18_15Mar~B01_1.mutationAnalysis.output.top
ma_cluconFrom4k-0-runallgivenref-AC222_D18_assembly.fa-222-18_14Mar~A01_1.mutationAnalysis.output.top
---- P222-D101
ma_cluconFrom4k-14-runallgivenref-X-222-101~C01_1.mutationAnalysis.output.top
ma_cluconFrom4k-15-runallgivenref-X-222-101_15Mar~C01_1.mutationAnalysis.output.top
ma_cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1.mutationAnalysis.output.top
---- P222-D105
ma_cluconFrom4k-3-runallgivenref-AC222_D105_assembly.fa-222-105_14Mar~B01_1.mutationAnalysis.output.top
ma_cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.mutationAnalysis.output.top
---- P9213-D59
ma_cluconFrom4k-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.mutationAnalysis.output.top
ma_cluconFrom4k-11-runallgivenref-9213_D59.fa-9213-59_15Mar~E01_1.mutationAnalysis.output.top
---- P9213-D165
ma_cluconFrom4k-12-runallgivenref-9213_D165.fa-9213-165_14Mar~F01_1.mutationAnalysis.output.top
ma_cluconFrom4k-13-runallgivenref-9213_D165.fa-9213-165_15Mar~H01_1.mutationAnalysis.output.top
Next
================================