goal: validate the large ~300-bp deletion observed in the HIV data.
ALIGNMENT RESULTS
Show IGV alignments for P222-D105 (strong deletion) and P9213-D59
(evidence of deletion but at lower abundance), P222-D101 (no large
deletion evidence by metrics)
Note, you might have to download the references and bam locally so IGV
can properly index them (the webserver is password protected).
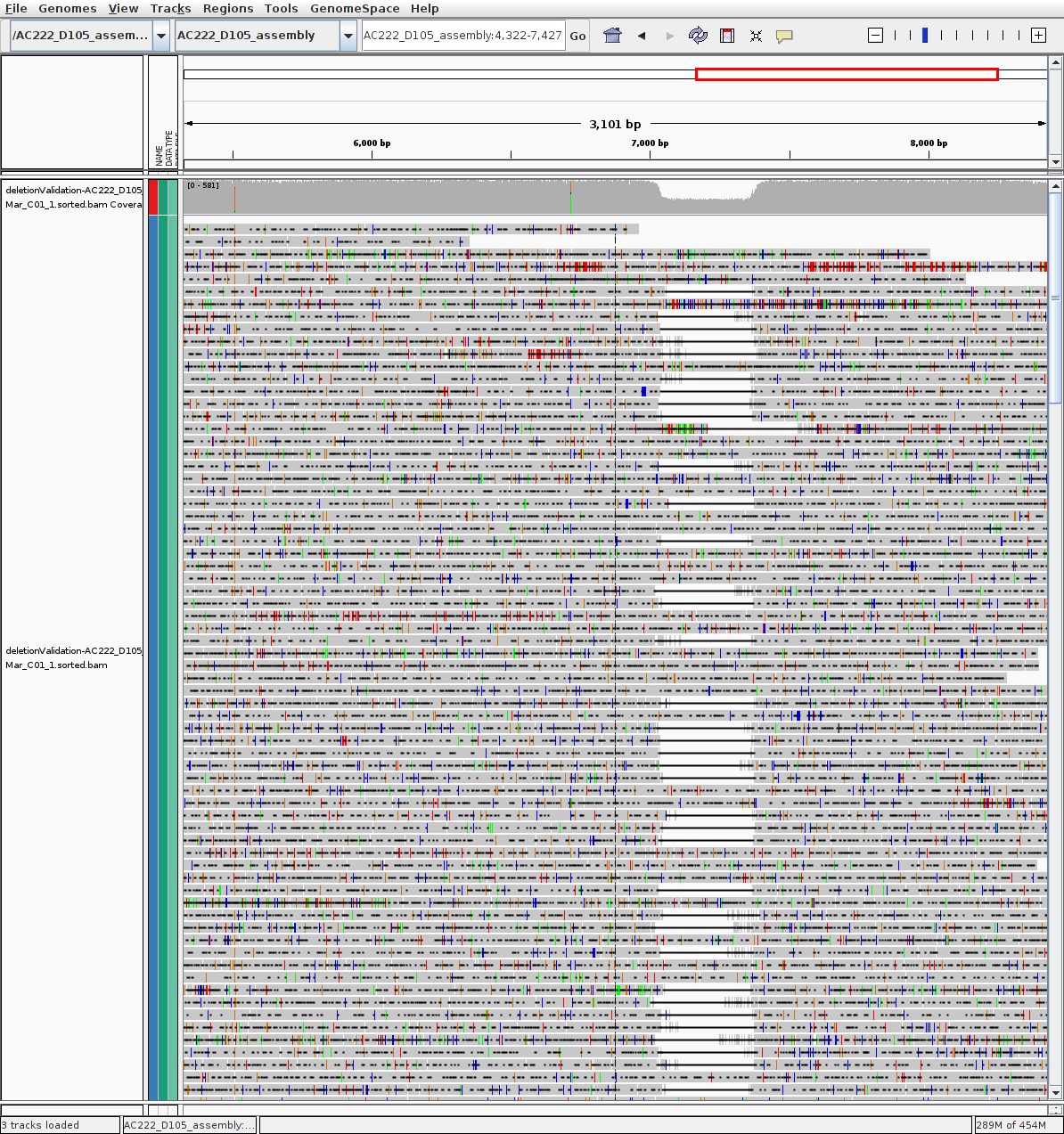
P222-D105 (strong deletion)
Given Reference: AC222_D105_assembly.fa
Given Reference: AC222_D105_assembly.fa.fai
BAM alignments: deletionValidation-AC222_D105_15Mar_C01_1.sorted.bam
BAM alignments: deletionValidation-AC222_D105_15Mar_C01_1.sorted.bam.bai
RAW reads >4k mapped: deletionValidation-clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.clustered.fasta
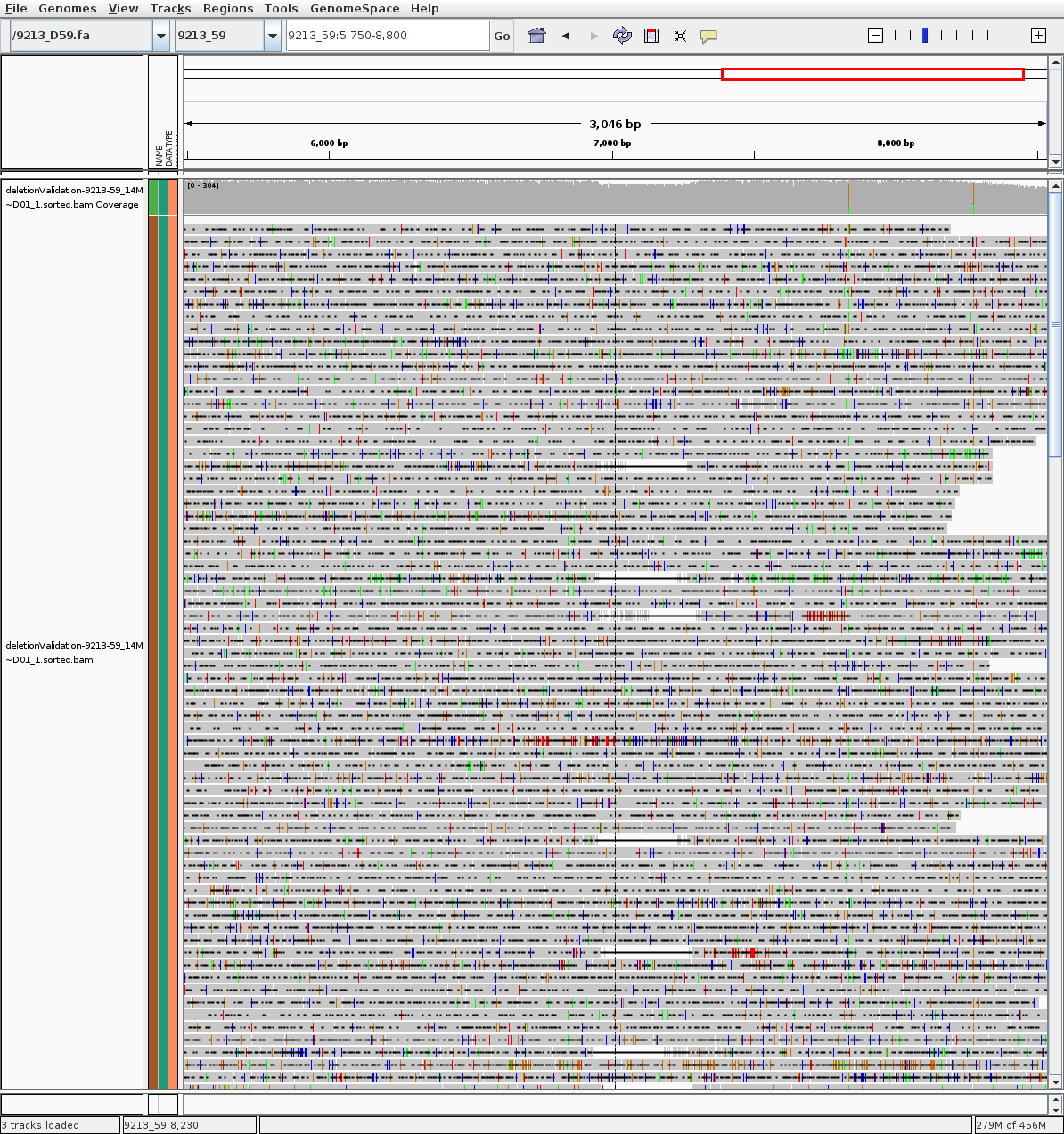
P9213-D59 (evidence of deletion)
Given Reference: 9213_D59.fa
Given Reference: 9213_D59.fa.fai
BAM alignments: deletionValidation-9213-59_14Mar~D01_1.sorted.bam
BAM alignments: deletionValidation-9213-59_14Mar~D01_1.sorted.bam.bai
RAW read >4k mapped: deletionValidation-clucon-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.clustered.fasta
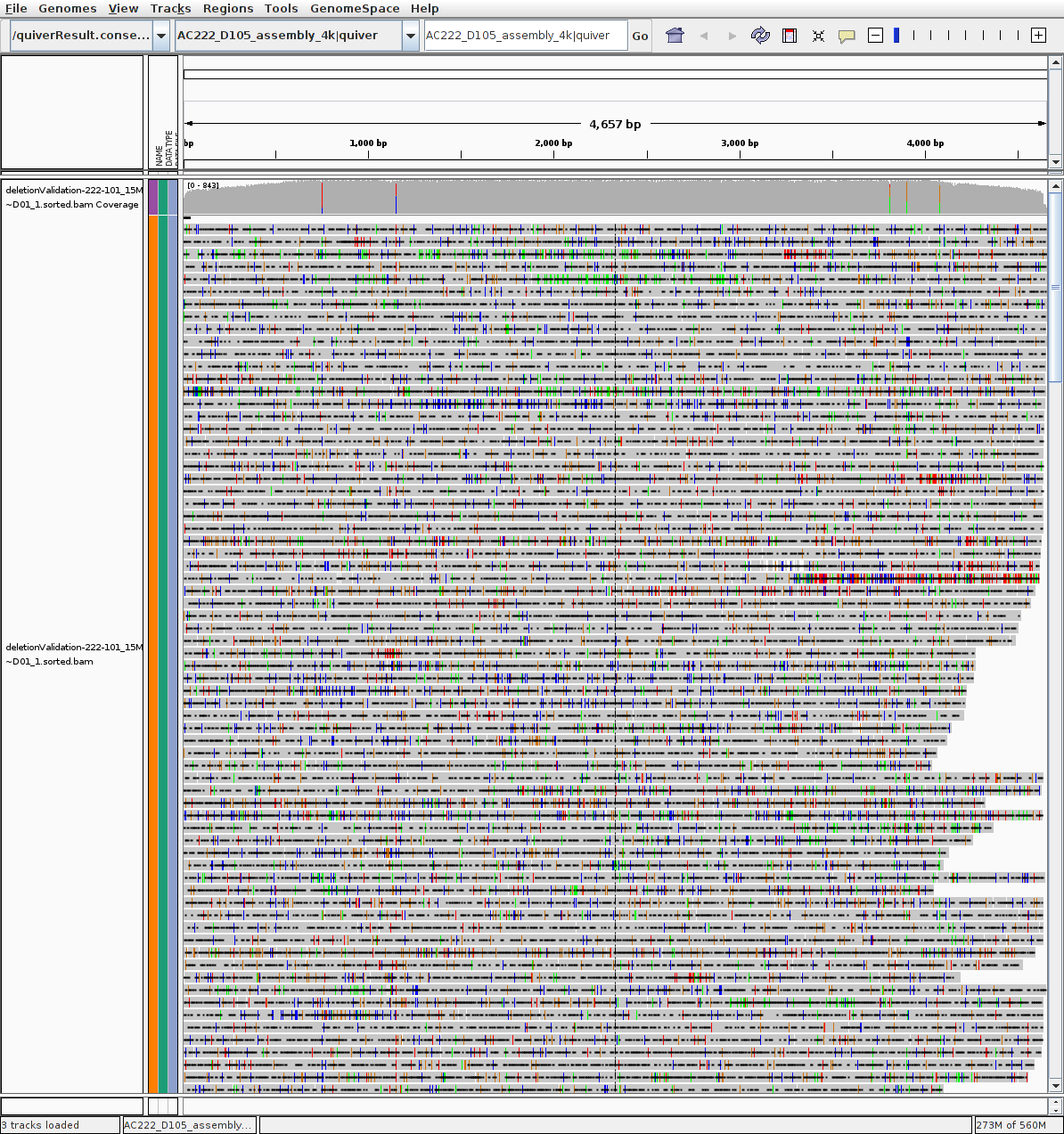
P222-D101 (no large deletion evidence)
Estimated eference: quiverResult.consensus.fasta
Estimated eference: quiverResult.consensus.fasta.fai
BAM alignments: deletionValidation-222-101_15Mar~D01_1.sorted.bam
BAM alignments: deletionValidation-222-101_15Mar~D01_1.sorted.bam.bai
RAW read >4k mapped: deletionValidation-cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1.clustered.fasta
METHODS:
Here is Rebecca's email Sept 2
>>>>>>>>>>>>>>>>
Hi Michael,
I am looking at AC222 D105, where you have a 300 bp deletion in Env in
your consensus reads relative to the reference genome that I
provided. However, I dont see the deletion in either the 454 reads or the Pac Bio reads when I look at an alignment of the raw reads to my reference. I use the program IGV to view the read alignment to the reference.
It is clear in the 454 reads that there is no deletion in Env relative
to the reference. In the alignment of the Pac Bio reads, I do see a
drop in coverage in that region, but it does not appear that there are
reads spanning that region that have missing bases.
In order to check the reads that you used, I selected reads longer
than 4000 bp and did the same alignment to my D105 reference. I get
about 50 reads. In these reads I also dont see a large deletion, in
fact these reads seem to nicely span the whole region.
Do you think this is a reasonable way to check for the deletion in the
reads? I wonder what could be going on in your consensus builder that
you get these deletions.
I also checked the 454 reads for sample 9213 D59, and I also see no
signs of such a large deletion.
I'll hold off on installing the software until we iron this out.
Thanks for any help with this.
Best,
Rebecca
--
Rebecca Batorsky, PhD
Research Fellow
Ragon Institute of MGH, MIT and Harvard
400 Technology Square | Room 761 | Cambridge MA 02139
Phone: 857.268.7015 Email: rbatorsky@partners.org
<<<<<<<<<<<<<<<<<
================================
GOAL: generate alignments of two time points in BAM format so that
Rebecca can see the deletion in IGV viewer.
TODO: Perhaps also give references for both deleted and non-deleted so
partners.org can run their pipeline and show that the deleted and
non-deleted alignments are truncations of each other (their aligner
doesn't want to follow the 300 base deletion and stops on one side,
giving a "half alignment".
Against their reference I have:
Initial Look:
clucon_222-105-15Mar-AC222_D105_assembly_fixed
Run all look:
clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/
ma_clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.mutationAnalysis.output.top
ma_clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.mutationAnalysis.output.png
Bases 7033-7382 strongly deleted. MSA 35165-36910
The recursion on the 4k sub-reference:
* drwxr-xr-x 2 mbrown domain_users 1237 2013-08-26 17:07 cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1
* drwxr-xr-x 2 mbrown domain_users 1436 2013-08-27 13:24 cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1-~_D1C1
* drwxr-xr-x 2 mbrown domain_users 1292 2013-08-27 13:17 cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1-~_D1C2
* drwxr-xr-x 2 mbrown domain_users 1292 2013-08-27 13:16 cluconFrom4k-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1-~_D1C3
In
clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/, I
have 590 reads that map >4k to their reference (+1 for the given reference).
The ids are in alignments.filterFull
265 265 17977 clustergroup_1.ids
202 202 13707 clustergroup_2.ids
74 74 5021 clustergroup_3.ids
49 49 3325 clustergroup_4.ids
590 590 40030 total
Let's create a fasta in the order of clustergroup_*.ids. If the
aligner puts them in the same order then the alignment should clearly
show the deletion. (IGV with SAM/BAM will change order... but only
look at the >4k mapped)
Pull the fasta in order of the clustering:
export SEYMOUR_HOME=/mnt/secondary/Smrtanalysis/opt/smrtanalysis; . $SEYMOUR_HOME/etc/setup.sh;
fastafetch --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta \
--index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta.index \
-F -q /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/clustergroup_1.ids \
> /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/deletionValidation-clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.clustered.fasta
fastafetch --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta \
--index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta.index \
-F -q /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/clustergroup_2.ids \
>> /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/deletionValidation-clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.clustered.fasta
fastafetch --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta \
--index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta.index \
-F -q /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/clustergroup_3.ids \
>> /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/deletionValidation-clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.clustered.fasta
fastafetch --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta \
--index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-105_15Mar/C01_1/Analysis_Results/m130316_025735_42183_c100458832550000001523065103201322_s1_p0.fasta.index \
-F -q /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1/clustergroup_4.ids \
>> /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/deletionValidation-clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.clustered.fasta
There are 590 sequences in that file.
----
Run pbalign.py to get a sam file:
pbalign.py deletionValidation-clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.clustered.fasta \
/mnt/data3/vol53/Share/CollaboratorData/partorg/data/assembly_fasta/AC222_D105_assembly.fa \
deletionValidation-AC222_D105_15Mar_C01_1.sam
# convert sam to input for IGV
samtools view -bS deletionValidation-AC222_D105_15Mar_C01_1.sam > deletionValidation-AC222_D105_15Mar_C01_1.bam
samtools sort deletionValidation-AC222_D105_15Mar_C01_1.bam deletionValidation-AC222_D105_15Mar_C01_1.sorted
samtools index deletionValidation-AC222_D105_15Mar_C01_1.sorted.bam
cp /mnt/data3/vol53/Share/CollaboratorData/partorg/data/assembly_fasta/AC222_D105_assembly.fa ./
>>>> Run IGV:
Reference: AC222_D105_assembly.fa
BAM alignments: deletionValidation-AC222_D105_15Mar_C01_1.sorted.bam
RAW reads >4k mapped: deletionValidation-clucon-4-runallgivenref-AC222_D105_assembly.fa-222-105_15Mar~C01_1.clustered.fasta
================================
Do the same for P9213-D59 in which the deletion should occur at lower
abundance.
cat clucon-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1/clustergroup_*.ids > /tmp/all.ids
fastaindex --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/9213-59_14Mar/D01_1/Analysis_Results/m130315_025009_42183_c100458702550000001523065103201383_s1_p0.fasta --index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/9213-59_14Mar/D01_1/Analysis_Results/m130315_025009_42183_c100458702550000001523065103201383_s1_p0.fasta.index
fastafetch --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/9213-59_14Mar/D01_1/Analysis_Results/m130315_025009_42183_c100458702550000001523065103201383_s1_p0.fasta \
--index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/9213-59_14Mar/D01_1/Analysis_Results/m130315_025009_42183_c100458702550000001523065103201383_s1_p0.fasta.index \
-F -q /tmp/all.ids \
> /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/deletionValidation-clucon-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.clustered.fasta
306 raw sequences
pbalign.py \
deletionValidation-clucon-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.clustered.fasta \
/mnt/data3/vol53/Share/CollaboratorData/partorg/data/assembly_fasta/9213_D59.fa \
deletionValidation-9213-59_14Mar~D01_1.sam
# convert sam to input for IGV: deletionValidation-9213-59_14Mar~D01_1
samtools view -bS $1.sam > $1.bam
samtools sort $1.bam $1.sorted
samtools index $1.sorted.bam
cp /mnt/data3/vol53/Share/CollaboratorData/partorg/data/assembly_fasta/9213_D59.fa ./
>>>> Run IGV:
Reference: 9213_D59.fa
BAM alignments: deletionValidation-9213-59_14Mar~D01_1.sorted.bam
RAW read >4k mapped: deletionValidation-clucon-10-runallgivenref-9213_D59.fa-9213-59_14Mar~D01_1.clustered.fasta
================================
Finally do P222-D101 (no observed deletion) on the estimated reference
because I don't have a given reference.
cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1:
cat cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1/clustergroup_*.ids > /tmp/all.ids
fastaindex --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-101_15Mar/D01_1/Analysis_Results/m130316_051506_42183_c100458832550000001523065103201323_s1_p0.fasta --index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-101_15Mar/D01_1/Analysis_Results/m130316_051506_42183_c100458832550000001523065103201323_s1_p0.fasta.index
fastafetch --fasta /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-101_15Mar/D01_1/Analysis_Results/m130316_051506_42183_c100458832550000001523065103201323_s1_p0.fasta --index /mnt/data3/vol53/Share/CollaboratorData/partorg/data/222-101_15Mar/D01_1/Analysis_Results/m130316_051506_42183_c100458832550000001523065103201323_s1_p0.fasta.index \
-F -q /tmp/all.ids \
> /home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/deletionValidation-cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1.clustered.fasta
850 raw sequences
pbalign.py \
deletionValidation-cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1.clustered.fasta \
/home/UNIXHOME/mbrown/mbrown/workspace2013Q3/partners-hiv/cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1/quiverResult.consensus.fasta \
deletionValidation-222-101_15Mar~D01_1.sam
# convert sam to input for IGV: deletionValidation-222-101_15Mar~D01_1
samtools view -bS $1.sam > $1.bam
samtools sort $1.bam $1.sorted
samtools index $1.sorted.bam
cp -i cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1/quiverResult.consensus.fasta quiverResult.consensus.fasta
>>>> Run IGV:
Reference: quiverResult.consensus.fasta
BAM alignments: deletionValidation-222-101_15Mar~D01_1.sorted.bam
RAW read >4k mapped: deletionValidation-cluconFrom4k-16-runallgivenref-X-222-101_15Mar~D01_1.clustered.fasta


