goal: latest figures, tables for BCR-ABL paper submission
================================================================
---- Highlight long read lengths.
We are doing 3kb CCS for HIV projects. Cover the entire kinase domain?
================================================================
---- Plot all variant positions in time series, not just the variable ones.
Here are all variants:
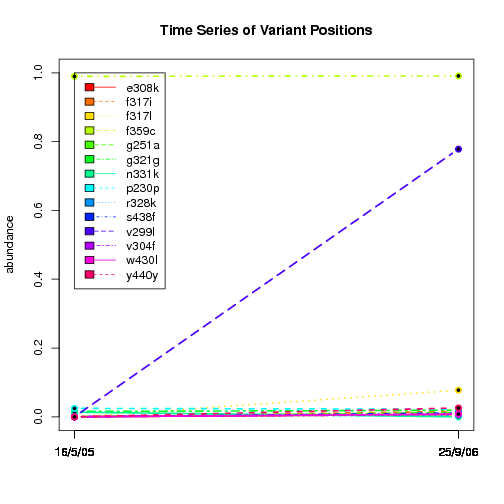
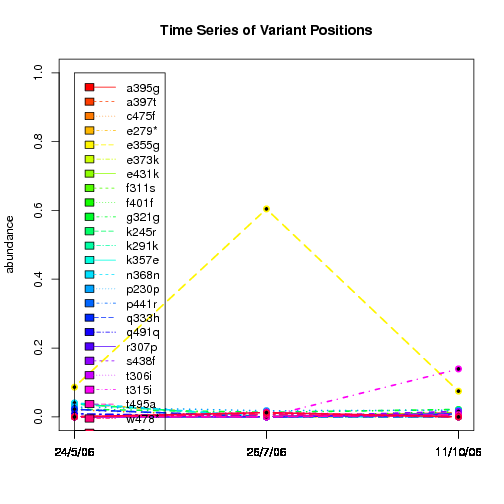
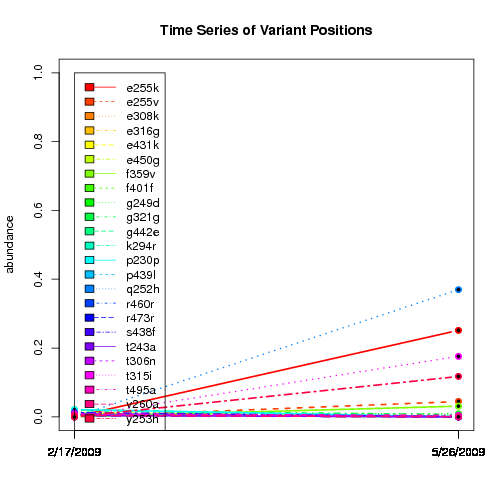
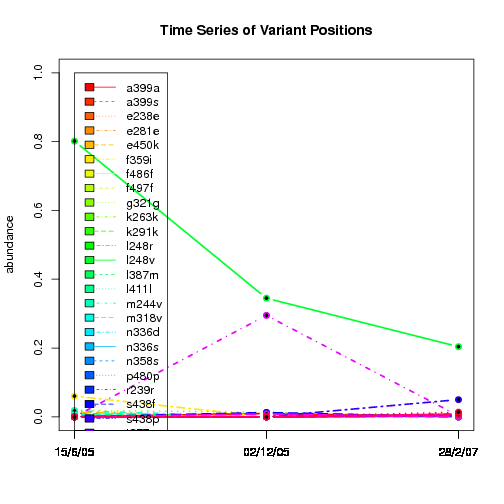
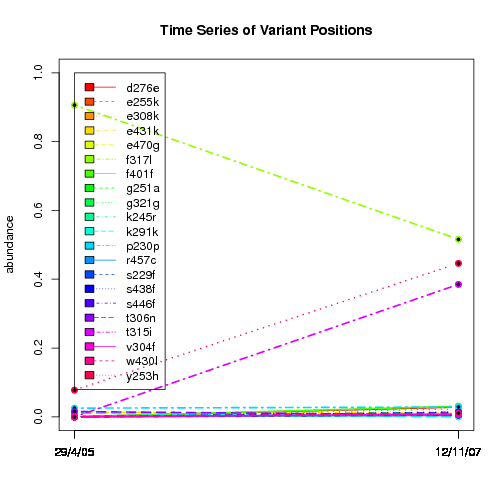
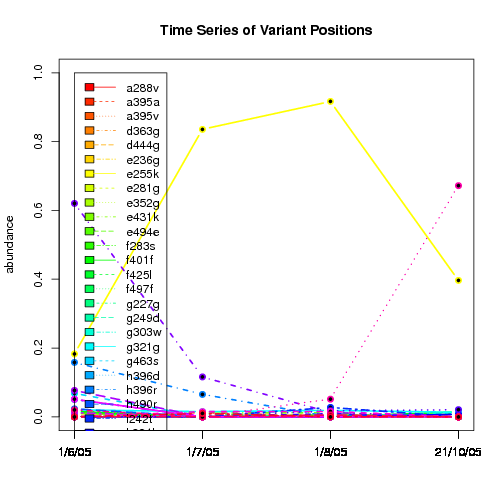
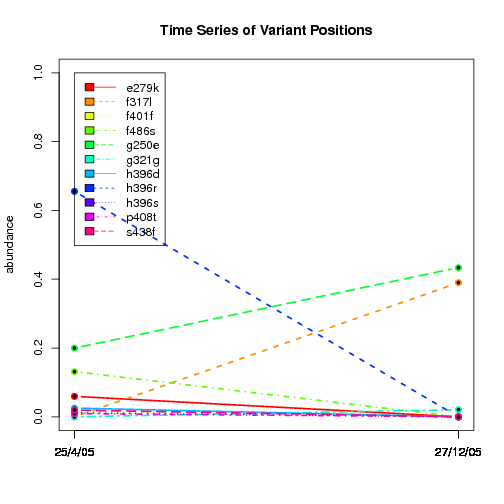
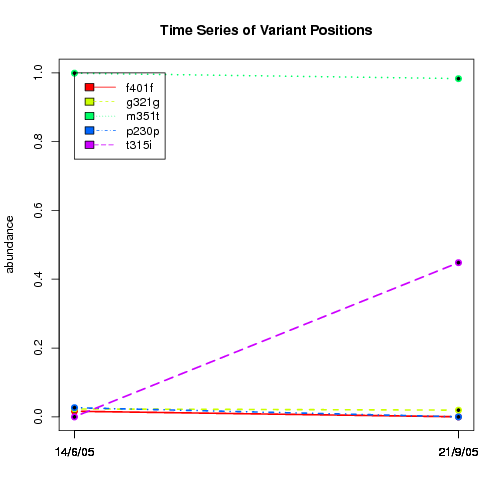
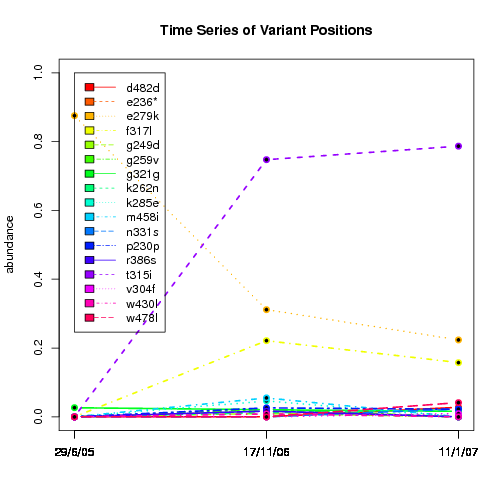
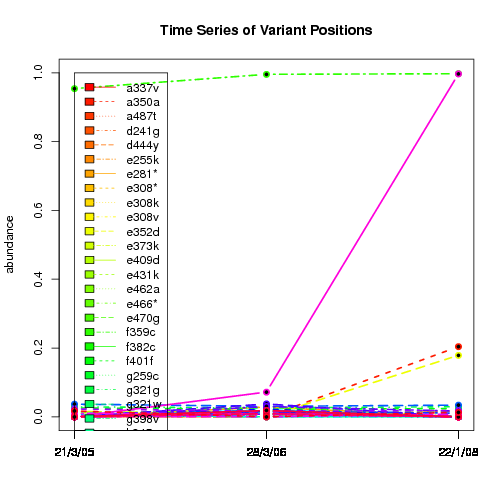
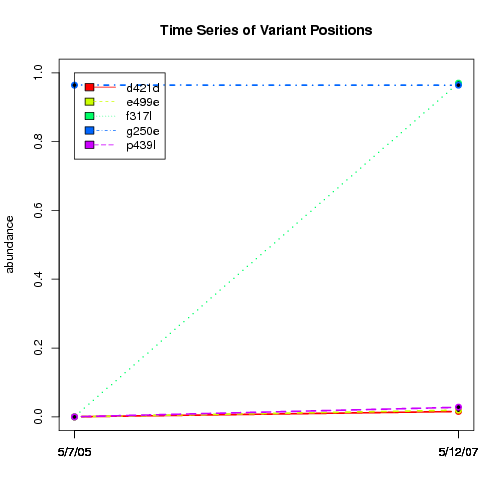
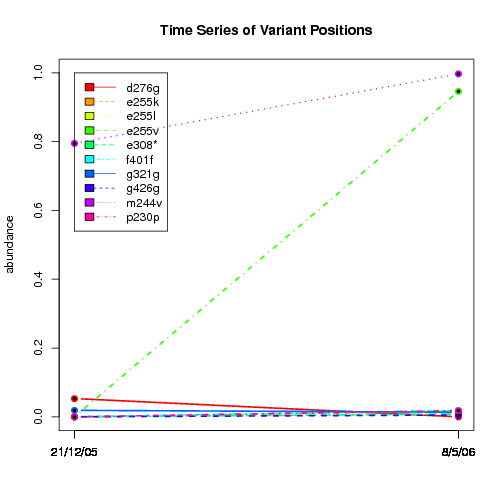
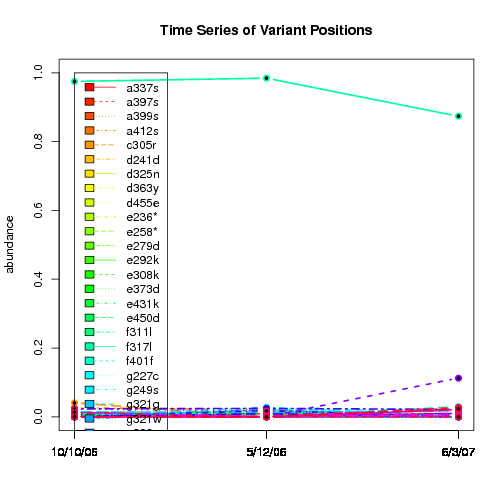 Some plots are too busy.
================================================================
Some plots are too busy.
================================================================
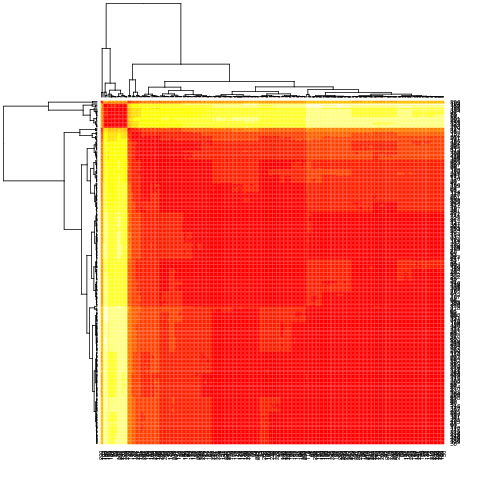
Clustering plots
Clustering plots for CSY time series:
2450177-0033.F1 21/3/05 2450177-0033.F2 28/3/06 2450177-0033.F3 22/1/08
CSY.21305 is simple (mostly f359c and wildtype), has large deletion in about 10% of reads
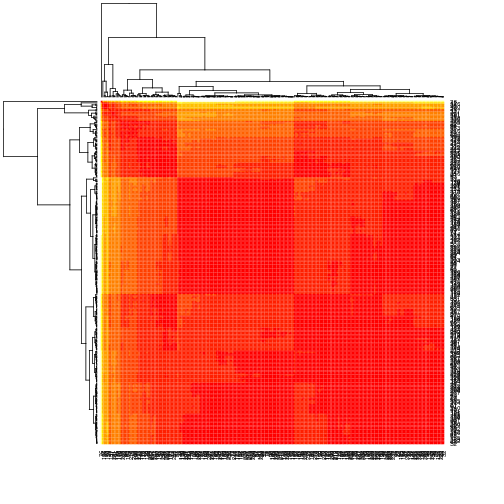
 CSY.28036 is simple (mostly f359c and wildtype, t315i+f359c at 1%), has large deletions in about 10% of reads
CSY.28036 is simple (mostly f359c and wildtype, t315i+f359c at 1%), has large deletions in about 10% of reads
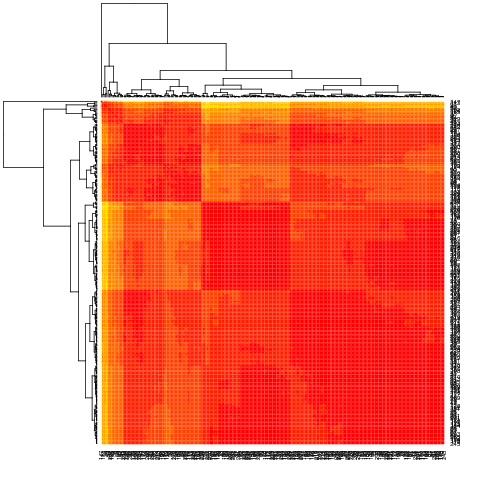
 CSY.22108 is complex with 8 compounds above 1% but no large deletions
CSY.22108 is complex with 8 compounds above 1% but no large deletions
 JLR 12/11/07 has most reads (81%) containing a 183 base consistent
deletion (61 amino acids) starting about halfway into the amplicon.
JLR 12/11/07 has most reads (81%) containing a 183 base consistent
deletion (61 amino acids) starting about halfway into the amplicon.
 Here are some 256-read multiple sequence alignments to show the
deletes. Note the aligner does not model large delete and tends to
fill in the delete with flanking sequence.
MSA CSY.21305
MSA CSY.28036
MSA CSY.22108
MSA JLR.121107
Note that large deletions are _not_ a PacBio error mode, so most
likely are not sequencing artifacts. I don't know whether PCR could
cause large consistent deletions.
================================================================
---- Cross-over PCR in compound mutations
Consider double compounds that might be caused by PCR cross-over.
First consider single cross over events with rates 13%-40%. Multiple
cross overs probably happen also.
I will consider breaking crosses: (A+B)X(None) = (A),(B) and building
crosses: (A)X(B) = (A+B)
Find a minor that is a compound of more abundant. Do the probablities
discount it away?
Here are all the discounted two component crosses (some are not found in both repeats):
discountBuild 2450177-0029.F2 AHP 6/3/07 q252h.cac,f317l.ttg 0.015522 q252h.cac 0.038806 f317l.ttg 0.52597 cross 0.760480050793
discountBreak 2450177-0044.F5 BHK 24/5/05 t315i.att,l387f.ttc 0.038298 none 0.278723 l387f.ttc 0.010638 cross 0.996577512811
discountBuild 2450177-0035.F7 BRM 21/12/05 m244v.gtg,d276g.ggc 0.013185 m244v.gtg 0.531947 d276g.ggc 0.024848 cross 0.997517059671
discountBuild 2450177-0026.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.032007 t315i.att 0.070069 h396r.cgt 0.472318 cross 0.967129328463
discountBuild 2450177-0027.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.028529 t315i.att 0.064565 h396r.cgt 0.459459 cross 0.961706675511
discountBuild 2450177-0047.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019909 f317i.atc 0.403982 w476c.tgt 0.055164 cross 0.893370652934
discountBuild 2450177-0046.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019769 f317i.atc 0.392391 w476c.tgt 0.054457 cross 0.925149569413
discountBreak 2450177-0036.F4 DWB 21/9/05 t315i.att,m351t.acg 0.236657 t315i.att 0.047331 none 0.246727 cross 0.810605688839
discountBuild 2450177-0052.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.013311 g250e.gag 0.059727 e255k.aag 0.385324 cross 0.578380872571
discountBuild 2450177-0053.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.017427 g250e.gag 0.059497 e255k.aag 0.368597 cross 0.794649779158
discountBuild 2450177-0021.F4 KM 5/7/05 g250e.gag,e255k.aag 0.011838 g250e.gag 0.057321 e255k.aag 0.507165 cross 0.407207063287
discountBuild 2450177-0020.F4 KM 5/7/05 g250e.gag,e255k.aag 0.014414 g250e.gag 0.064702 e255k.aag 0.529148 cross 0.421007326292
discountBreak 2450177-0034.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.181511 none 0.409582 y253f.ttc 0.046559 cross 0.626267438624
discountBreak 2450177-0035.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.169377 none 0.415031 y253f.ttc 0.055524 cross 0.789852091323
discountBuild 2450177-0030.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.012014 l248v.gtg 0.073145 v299l.ttg 0.192226 cross 0.854458264048
discountBuild 2450177-0031.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.011028 l248v.gtg 0.076485 v299l.ttg 0.176805 cross 0.815503715054
discountBuild 2450177-0034.F1 NEF 25/9/06 v299l.ctg,f317l.ctc 0.01169 v299l.ctg 0.388818 f317l.ctc 0.03202 cross 0.938959416227
For example in the first line, in one of the AHP 6/3/07 runs: you can
explain q252h+f317l compound at 1.5% by taking q252h at 3.9% along
with f317l at 52.6% and crossing them at a rate of 76%. (1.5% =
3.9%*52.6%*76%).
I can consider more than two components with more logic, and multiple
crosses.
================================================================
Here are some 256-read multiple sequence alignments to show the
deletes. Note the aligner does not model large delete and tends to
fill in the delete with flanking sequence.
MSA CSY.21305
MSA CSY.28036
MSA CSY.22108
MSA JLR.121107
Note that large deletions are _not_ a PacBio error mode, so most
likely are not sequencing artifacts. I don't know whether PCR could
cause large consistent deletions.
================================================================
---- Cross-over PCR in compound mutations
Consider double compounds that might be caused by PCR cross-over.
First consider single cross over events with rates 13%-40%. Multiple
cross overs probably happen also.
I will consider breaking crosses: (A+B)X(None) = (A),(B) and building
crosses: (A)X(B) = (A+B)
Find a minor that is a compound of more abundant. Do the probablities
discount it away?
Here are all the discounted two component crosses (some are not found in both repeats):
discountBuild 2450177-0029.F2 AHP 6/3/07 q252h.cac,f317l.ttg 0.015522 q252h.cac 0.038806 f317l.ttg 0.52597 cross 0.760480050793
discountBreak 2450177-0044.F5 BHK 24/5/05 t315i.att,l387f.ttc 0.038298 none 0.278723 l387f.ttc 0.010638 cross 0.996577512811
discountBuild 2450177-0035.F7 BRM 21/12/05 m244v.gtg,d276g.ggc 0.013185 m244v.gtg 0.531947 d276g.ggc 0.024848 cross 0.997517059671
discountBuild 2450177-0026.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.032007 t315i.att 0.070069 h396r.cgt 0.472318 cross 0.967129328463
discountBuild 2450177-0027.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.028529 t315i.att 0.064565 h396r.cgt 0.459459 cross 0.961706675511
discountBuild 2450177-0047.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019909 f317i.atc 0.403982 w476c.tgt 0.055164 cross 0.893370652934
discountBuild 2450177-0046.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019769 f317i.atc 0.392391 w476c.tgt 0.054457 cross 0.925149569413
discountBreak 2450177-0036.F4 DWB 21/9/05 t315i.att,m351t.acg 0.236657 t315i.att 0.047331 none 0.246727 cross 0.810605688839
discountBuild 2450177-0052.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.013311 g250e.gag 0.059727 e255k.aag 0.385324 cross 0.578380872571
discountBuild 2450177-0053.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.017427 g250e.gag 0.059497 e255k.aag 0.368597 cross 0.794649779158
discountBuild 2450177-0021.F4 KM 5/7/05 g250e.gag,e255k.aag 0.011838 g250e.gag 0.057321 e255k.aag 0.507165 cross 0.407207063287
discountBuild 2450177-0020.F4 KM 5/7/05 g250e.gag,e255k.aag 0.014414 g250e.gag 0.064702 e255k.aag 0.529148 cross 0.421007326292
discountBreak 2450177-0034.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.181511 none 0.409582 y253f.ttc 0.046559 cross 0.626267438624
discountBreak 2450177-0035.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.169377 none 0.415031 y253f.ttc 0.055524 cross 0.789852091323
discountBuild 2450177-0030.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.012014 l248v.gtg 0.073145 v299l.ttg 0.192226 cross 0.854458264048
discountBuild 2450177-0031.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.011028 l248v.gtg 0.076485 v299l.ttg 0.176805 cross 0.815503715054
discountBuild 2450177-0034.F1 NEF 25/9/06 v299l.ctg,f317l.ctc 0.01169 v299l.ctg 0.388818 f317l.ctc 0.03202 cross 0.938959416227
For example in the first line, in one of the AHP 6/3/07 runs: you can
explain q252h+f317l compound at 1.5% by taking q252h at 3.9% along
with f317l at 52.6% and crossing them at a rate of 76%. (1.5% =
3.9%*52.6%*76%).
I can consider more than two components with more logic, and multiple
crosses.
================================================================












 Some plots are too busy.
================================================================
Some plots are too busy.
================================================================
 CSY.28036 is simple (mostly f359c and wildtype, t315i+f359c at 1%), has large deletions in about 10% of reads
CSY.28036 is simple (mostly f359c and wildtype, t315i+f359c at 1%), has large deletions in about 10% of reads
 CSY.22108 is complex with 8 compounds above 1% but no large deletions
CSY.22108 is complex with 8 compounds above 1% but no large deletions
 JLR 12/11/07 has most reads (81%) containing a 183 base consistent
deletion (61 amino acids) starting about halfway into the amplicon.
JLR 12/11/07 has most reads (81%) containing a 183 base consistent
deletion (61 amino acids) starting about halfway into the amplicon.
 Here are some 256-read multiple sequence alignments to show the
deletes. Note the aligner does not model large delete and tends to
fill in the delete with flanking sequence.
MSA CSY.21305
MSA CSY.28036
MSA CSY.22108
MSA JLR.121107
Note that large deletions are _not_ a PacBio error mode, so most
likely are not sequencing artifacts. I don't know whether PCR could
cause large consistent deletions.
================================================================
---- Cross-over PCR in compound mutations
Consider double compounds that might be caused by PCR cross-over.
First consider single cross over events with rates 13%-40%. Multiple
cross overs probably happen also.
I will consider breaking crosses: (A+B)X(None) = (A),(B) and building
crosses: (A)X(B) = (A+B)
Find a minor that is a compound of more abundant. Do the probablities
discount it away?
Here are all the discounted two component crosses (some are not found in both repeats):
discountBuild 2450177-0029.F2 AHP 6/3/07 q252h.cac,f317l.ttg 0.015522 q252h.cac 0.038806 f317l.ttg 0.52597 cross 0.760480050793
discountBreak 2450177-0044.F5 BHK 24/5/05 t315i.att,l387f.ttc 0.038298 none 0.278723 l387f.ttc 0.010638 cross 0.996577512811
discountBuild 2450177-0035.F7 BRM 21/12/05 m244v.gtg,d276g.ggc 0.013185 m244v.gtg 0.531947 d276g.ggc 0.024848 cross 0.997517059671
discountBuild 2450177-0026.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.032007 t315i.att 0.070069 h396r.cgt 0.472318 cross 0.967129328463
discountBuild 2450177-0027.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.028529 t315i.att 0.064565 h396r.cgt 0.459459 cross 0.961706675511
discountBuild 2450177-0047.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019909 f317i.atc 0.403982 w476c.tgt 0.055164 cross 0.893370652934
discountBuild 2450177-0046.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019769 f317i.atc 0.392391 w476c.tgt 0.054457 cross 0.925149569413
discountBreak 2450177-0036.F4 DWB 21/9/05 t315i.att,m351t.acg 0.236657 t315i.att 0.047331 none 0.246727 cross 0.810605688839
discountBuild 2450177-0052.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.013311 g250e.gag 0.059727 e255k.aag 0.385324 cross 0.578380872571
discountBuild 2450177-0053.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.017427 g250e.gag 0.059497 e255k.aag 0.368597 cross 0.794649779158
discountBuild 2450177-0021.F4 KM 5/7/05 g250e.gag,e255k.aag 0.011838 g250e.gag 0.057321 e255k.aag 0.507165 cross 0.407207063287
discountBuild 2450177-0020.F4 KM 5/7/05 g250e.gag,e255k.aag 0.014414 g250e.gag 0.064702 e255k.aag 0.529148 cross 0.421007326292
discountBreak 2450177-0034.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.181511 none 0.409582 y253f.ttc 0.046559 cross 0.626267438624
discountBreak 2450177-0035.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.169377 none 0.415031 y253f.ttc 0.055524 cross 0.789852091323
discountBuild 2450177-0030.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.012014 l248v.gtg 0.073145 v299l.ttg 0.192226 cross 0.854458264048
discountBuild 2450177-0031.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.011028 l248v.gtg 0.076485 v299l.ttg 0.176805 cross 0.815503715054
discountBuild 2450177-0034.F1 NEF 25/9/06 v299l.ctg,f317l.ctc 0.01169 v299l.ctg 0.388818 f317l.ctc 0.03202 cross 0.938959416227
For example in the first line, in one of the AHP 6/3/07 runs: you can
explain q252h+f317l compound at 1.5% by taking q252h at 3.9% along
with f317l at 52.6% and crossing them at a rate of 76%. (1.5% =
3.9%*52.6%*76%).
I can consider more than two components with more logic, and multiple
crosses.
================================================================
Here are some 256-read multiple sequence alignments to show the
deletes. Note the aligner does not model large delete and tends to
fill in the delete with flanking sequence.
MSA CSY.21305
MSA CSY.28036
MSA CSY.22108
MSA JLR.121107
Note that large deletions are _not_ a PacBio error mode, so most
likely are not sequencing artifacts. I don't know whether PCR could
cause large consistent deletions.
================================================================
---- Cross-over PCR in compound mutations
Consider double compounds that might be caused by PCR cross-over.
First consider single cross over events with rates 13%-40%. Multiple
cross overs probably happen also.
I will consider breaking crosses: (A+B)X(None) = (A),(B) and building
crosses: (A)X(B) = (A+B)
Find a minor that is a compound of more abundant. Do the probablities
discount it away?
Here are all the discounted two component crosses (some are not found in both repeats):
discountBuild 2450177-0029.F2 AHP 6/3/07 q252h.cac,f317l.ttg 0.015522 q252h.cac 0.038806 f317l.ttg 0.52597 cross 0.760480050793
discountBreak 2450177-0044.F5 BHK 24/5/05 t315i.att,l387f.ttc 0.038298 none 0.278723 l387f.ttc 0.010638 cross 0.996577512811
discountBuild 2450177-0035.F7 BRM 21/12/05 m244v.gtg,d276g.ggc 0.013185 m244v.gtg 0.531947 d276g.ggc 0.024848 cross 0.997517059671
discountBuild 2450177-0026.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.032007 t315i.att 0.070069 h396r.cgt 0.472318 cross 0.967129328463
discountBuild 2450177-0027.F4 CSC 26/4/05 t315i.att,h396r.cgt 0.028529 t315i.att 0.064565 h396r.cgt 0.459459 cross 0.961706675511
discountBuild 2450177-0047.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019909 f317i.atc 0.403982 w476c.tgt 0.055164 cross 0.893370652934
discountBuild 2450177-0046.F3 DMJ 12/7/06 f317i.atc,w476c.tgt 0.019769 f317i.atc 0.392391 w476c.tgt 0.054457 cross 0.925149569413
discountBreak 2450177-0036.F4 DWB 21/9/05 t315i.att,m351t.acg 0.236657 t315i.att 0.047331 none 0.246727 cross 0.810605688839
discountBuild 2450177-0052.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.013311 g250e.gag 0.059727 e255k.aag 0.385324 cross 0.578380872571
discountBuild 2450177-0053.F0 EAD 5/1/06 g250e.gag,e255k.aag 0.017427 g250e.gag 0.059497 e255k.aag 0.368597 cross 0.794649779158
discountBuild 2450177-0021.F4 KM 5/7/05 g250e.gag,e255k.aag 0.011838 g250e.gag 0.057321 e255k.aag 0.507165 cross 0.407207063287
discountBuild 2450177-0020.F4 KM 5/7/05 g250e.gag,e255k.aag 0.014414 g250e.gag 0.064702 e255k.aag 0.529148 cross 0.421007326292
discountBreak 2450177-0034.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.181511 none 0.409582 y253f.ttc 0.046559 cross 0.626267438624
discountBreak 2450177-0035.F2 LYS 27/5/05 t240a.gcg,y253f.ttc 0.169377 none 0.415031 y253f.ttc 0.055524 cross 0.789852091323
discountBuild 2450177-0030.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.012014 l248v.gtg 0.073145 v299l.ttg 0.192226 cross 0.854458264048
discountBuild 2450177-0031.F3 MDL 02/12/05 l248v.gtg,v299l.ttg 0.011028 l248v.gtg 0.076485 v299l.ttg 0.176805 cross 0.815503715054
discountBuild 2450177-0034.F1 NEF 25/9/06 v299l.ctg,f317l.ctc 0.01169 v299l.ctg 0.388818 f317l.ctc 0.03202 cross 0.938959416227
For example in the first line, in one of the AHP 6/3/07 runs: you can
explain q252h+f317l compound at 1.5% by taking q252h at 3.9% along
with f317l at 52.6% and crossing them at a rate of 76%. (1.5% =
3.9%*52.6%*76%).
I can consider more than two components with more logic, and multiple
crosses.
================================================================